Astroglial-Mediated Remodeling of the Interhemispheric Midline Is Required for the Formation of the Corpus Callosum
- PMID: 27732850
- PMCID: PMC5094913
- DOI: 10.1016/j.celrep.2016.09.033
Astroglial-Mediated Remodeling of the Interhemispheric Midline Is Required for the Formation of the Corpus Callosum
Abstract
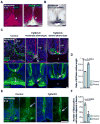
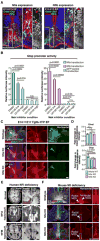
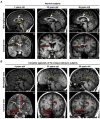
The corpus callosum is the major axon tract that connects and integrates neural activity between the two cerebral hemispheres. Although ∼1:4,000 children are born with developmental absence of the corpus callosum, the primary etiology of this condition remains unknown. Here, we demonstrate that midline crossing of callosal axons is dependent upon the prior remodeling and degradation of the intervening interhemispheric fissure. This remodeling event is initiated by astroglia on either side of the interhemispheric fissure, which intercalate with one another and degrade the intervening leptomeninges. Callosal axons then preferentially extend over these specialized astroglial cells to cross the midline. A key regulatory step in interhemispheric remodeling is the differentiation of these astroglia from radial glia, which is initiated by Fgf8 signaling to downstream Nfi transcription factors. Crucially, our findings from human neuroimaging studies reveal that developmental defects in interhemispheric remodeling are likely to be a primary etiology underlying human callosal agenesis.
Keywords: Fgf8; Nfia; Nfib; astrocyte; callosal agenesis; interhemispheric fissure.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Bagri A, Marin O, Plump AS, Mak J, Pleasure SJ, Rubenstein JL, Tessier-Lavigne M. Slit proteins prevent midline crossing and determine the dorsoventral position of major axonal pathways in the mammalian forebrain. Neuron. 2002;33:233–248. - PubMed
-
- Bremer C, Tung CH, Weissleder R. In vivo molecular target assessment of matrix metalloproteinase inhibition. Nature medicine. 2001;7:743–748. - PubMed
-
- Chen CP, Su YN, Chen YY, Chern SR, Liu YP, Wu PC, Lee CC, Chen YT, Wang W. Chromosome 1p32-p31 deletion syndrome: prenatal diagnosis by array comparative genomic hybridization using uncultured amniocytes and association with NFIA haploinsufficiency, ventriculomegaly, corpus callosum hypogenesis, abnormal external genitalia, and intrauterine growth restriction. Taiwanese journal of obstetrics & gynecology. 2011;50:345–352. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

