CNARA: reliability assessment for genomic copy number profiles
- PMID: 27733115
- PMCID: PMC5062840
- DOI: 10.1186/s12864-016-3074-7
CNARA: reliability assessment for genomic copy number profiles
Abstract
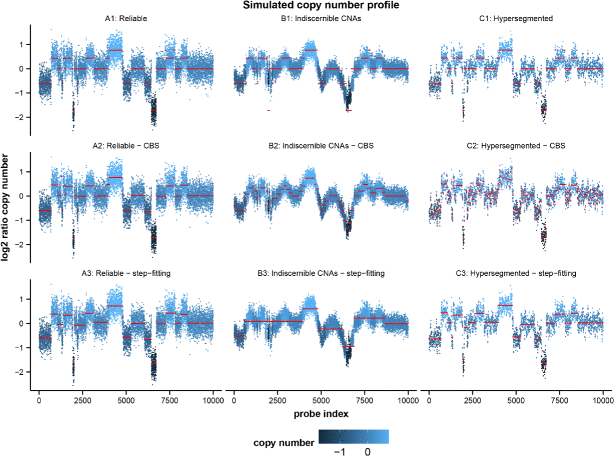
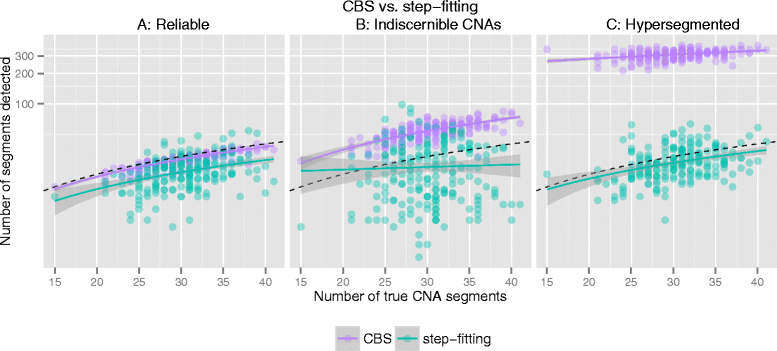
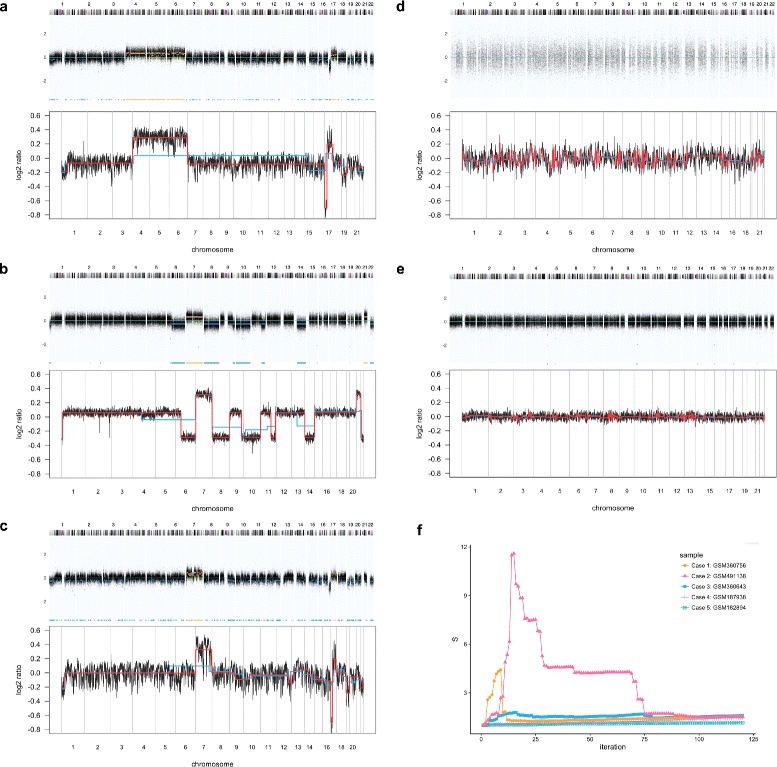
Background: DNA copy number profiles from microarray and sequencing experiments sometimes contain wave artefacts which may be introduced during sample preparation and cannot be removed completely by existing preprocessing methods. Besides, large derivative log ratio spread (DLRS) of the probes correlating with poor DNA quality is sometimes observed in genome screening experiments and may lead to unreliable copy number profiles. Depending on the extent of these artefacts and the resulting misidentification of copy number alterations/variations (CNA/CNV), it may be desirable to exclude such samples from analyses or to adapt the downstream data analysis strategy accordingly.
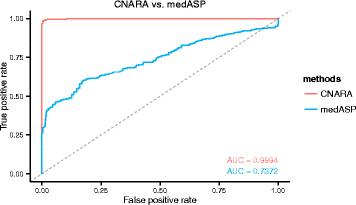
Results: Here, we propose a method to distinguish reliable genomic copy number profiles from those containing heavy wave artefacts and/or large DLRS. We define four features that adequately summarize the copy number profiles for reliability assessment, and train a classifier on a dataset of 1522 copy number profiles from various microarray platforms. The method can be applied to predict the reliability of copy number profiles irrespective of the underlying microarray platform and may be adapted for those sequencing platforms from which copy number estimates could be computed as a piecewise constant signal. Further details can be found at https://github.com/baudisgroup/CNARA .
Conclusions: We have developed a method for the assessment of genomic copy number profiling data, and suggest to apply the method in addition to and after other state-of-the-art noise correction and quality control procedures. CNARA could be instrumental in improving the assessment of data used for genomic data mining experiments and support the reliable functional attribution of copy number aberrations especially in cancer research.
Keywords: CNA; Copy number profile; Reliability assessment.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

