Proinflammatory cytokines decrease the expression of genes critical for RPE function
- PMID: 27733811
- PMCID: PMC5055142
Proinflammatory cytokines decrease the expression of genes critical for RPE function
Abstract
Purpose: Proinflammatory cytokines interferon gamma (IFN-γ), tumor necrosis factor alpha (TNF-α), and interleukin-1 beta (IL-1β) secreted by infiltrating lymphocytes or macrophages may play a role in triggering RPE dysfunction associated with age-related macular degeneration (AMD). Binding of these proinflammatory cytokines to their specific receptors residing on the RPE cell surface can activate signaling pathways that, in turn, may dysregulate cellular gene expression. The purpose of the present study was to investigate whether IFN-γ, TNF-α, and IL-1β have an adverse effect on the expression of genes essential for RPE function, employing the RPE cell line ARPE-19 as a model system.
Methods: ARPE-19 cells were cultured for 3-4 months until they exhibited epithelial morphology and expressed mRNAs for visual cycle genes. The differentiated cells were treated with IFN-γ, TNF-α, and/or IL-1β, and gene expression was analyzed with real-time PCR analysis. Western immunoblotting was employed for the detection of proteins.
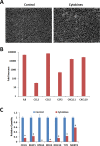
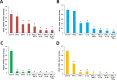
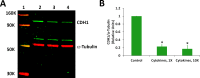
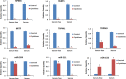
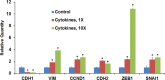
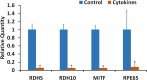
Results: Proinflammatory cytokines (IFN-γ + TNF-α + IL-1β) greatly increased the expression of chemokines and cytokines in cultured ARPE-19 cells that exhibited RPE characteristics. However, this response was accompanied by markedly decreased expression of genes important for RPE function, such as CDH1, RPE65, RDH5, RDH10, TYR, and MERTK. This was associated with decreased expression of the genes MITF, TRPM1, and TRPM3, as well as microRNAs miR-204 and miR-211, which are known to regulate RPE-specific gene expression. The decreased expression of the epithelial marker gene CDH1 was associated with increased expression of mesenchymal marker genes (CDH2, VIM, and CCND1) and epithelial-mesenchymal transition (EMT) promoting transcription factor genes (ZEB1 and SNAI1).
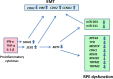
Conclusions: RPE cells exposed to proinflammatory cytokines IFN-γ, TNF-α, and IL-1β showed decreased expression of key genes involved in the visual cycle, epithelial morphology, and phagocytosis. This adverse effect of proinflammatory cytokines, which could be secreted by infiltrating lymphocytes or macrophages, on the expression of genes indispensable for RPE function may contribute to the RPE dysfunction implicated in AMD pathology.
Figures

References
-
- Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005;85:845–81. - PubMed
-
- Buschini E, Piras A, Nuzzi R, Vercelli A. Age related macular degeneration and drusen: neuroinflammation in the retina. Prog Neurobiol. 2011;95:14–25. - PubMed
-
- Cousins SW, Espinosa-Heidmann DG, Csaky KG. Monocyte activation in patients with age-related macular degeneration: a biomarker of risk for choroidal neovascularization? Arch Ophthalmol. 2004;122:1013–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

