Enterococcal Metabolite Cues Facilitate Interspecies Niche Modulation and Polymicrobial Infection
- PMID: 27736645
- PMCID: PMC5076562
- DOI: 10.1016/j.chom.2016.09.004
Enterococcal Metabolite Cues Facilitate Interspecies Niche Modulation and Polymicrobial Infection
Abstract
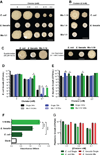
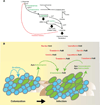
Enterococcus faecalis is frequently associated with polymicrobial infections of the urinary tract, indwelling catheters, and surgical wound sites. E. faecalis co-exists with Escherichia coli and other pathogens in wound infections, but mechanisms that govern polymicrobial colonization and pathogenesis are poorly defined. During infection, bacteria must overcome multiple host defenses, including nutrient iron limitation, to persist and cause disease. In this study, we investigated the contribution of E. faecalis to mixed-species infection when iron availability is restricted. We show that E. faecalis significantly augments E. coli biofilm growth and survival in vitro and in vivo by exporting L-ornithine. This metabolic cue facilitates E. coli biosynthesis of the enterobactin siderophore, allowing E. coli growth and biofilm formation in iron-limiting conditions that would otherwise restrict its growth. Thus, E. faecalis modulates its local environment by contributing growth-promoting cues that allow co-infecting organisms to overcome iron limitation and promotes polymicrobial infections.
Keywords: Enterococcus faecalis; Escherichia coli; iron; nutritional immunity; polymicrobial infection; wound infection.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures



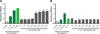
Comment in
-
Enterococcus faecalis: E. coli's Siderophore-Inducing Sidekick.Cell Host Microbe. 2016 Oct 12;20(4):411-412. doi: 10.1016/j.chom.2016.09.018. Cell Host Microbe. 2016. PMID: 27736638 Free PMC article.
References
-
- Archibald F. Manganese: its acquisition by and function in the lactic acid bacteria. Crit Rev Microbiol. 1986;13:63–109. - PubMed
-
- Browne AC, Vearncombe M, Sibbald RG. High bacterial load in asymptomatic diabetic patients with neurotrophic ulcers retards wound healing after application of Dermagraft. Ostomy Wound Manage. 2001;47:44–49. - PubMed
-
- Bruyneel B, Vandewoestyne M, Verstraete W. Lactic-Acid Bacteria - Microorganisms able to grow in the absence of available iron and copper. Biotechnology Letters. 1989;11:401–406.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

