Rheumatoid arthritis: identifying and characterising polymorphisms using rat models
- PMID: 27736747
- PMCID: PMC5087835
- DOI: 10.1242/dmm.026435
Rheumatoid arthritis: identifying and characterising polymorphisms using rat models
Abstract
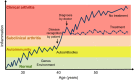
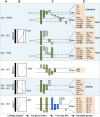
Rheumatoid arthritis is a chronic inflammatory joint disorder characterised by erosive inflammation of the articular cartilage and by destruction of the synovial joints. It is regulated by both genetic and environmental factors, and, currently, there is no preventative treatment or cure for this disease. Genome-wide association studies have identified ∼100 new loci associated with rheumatoid arthritis, in addition to the already known locus within the major histocompatibility complex II region. However, together, these loci account for only a modest fraction of the genetic variance associated with this disease and very little is known about the pathogenic roles of most of the risk loci identified. Here, we discuss how rat models of rheumatoid arthritis are being used to detect quantitative trait loci that regulate different arthritic traits by genetic linkage analysis and to positionally clone the underlying causative genes using congenic strains. By isolating specific loci on a fixed genetic background, congenic strains overcome the challenges of genetic heterogeneity and environmental interactions associated with human studies. Most importantly, congenic strains allow functional experimental studies be performed to investigate the pathological consequences of natural genetic polymorphisms, as illustrated by the discovery of several major disease genes that contribute to arthritis in rats. We discuss how these advances have provided new biological insights into arthritis in humans.
Keywords: Chronic inflammation; Congenic mapping; Genetics; Rat models; Rheumatoid arthritis; Susceptibility genes.
© 2016. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures


References
-
- Ahlqvist E., Ekman D., Lindvall T., Popovic M., Förster M., Hultqvist M., Klaczkowska D., Teneva I., Johannesson M., Flint J. et al. (2011). High-resolution mapping of a complex disease, a model for rheumatoid arthritis, using heterogeneous stock mice. Hum. Mol. Genet. 20, 3031-3041. 10.1093/hmg/ddr206 - DOI - PubMed
-
- Aho K., Palosuo T., Heliövaara M., Knekt P., Alha P. and von Essen R. (2000). Antifilaggrin antibodies within “normal” range predict rheumatoid arthritis in a linear fashion. J. Rheumatol. 27, 2743-2746. - PubMed
-
- Aletaha D., Neogi T., Silman A. J., Funovits J., Felson D. T., Bingham C. O., Birnbaum N. S., Burmester G. R., Bykerk V. P., Cohen M. D. et al. (2010). 2010 Rheumatoid arthritis classification criteria: an American college of rheumatology/European league against rheumatism collaborative initiative. Arthritis. Rheum. 62, 2569-2581. 10.1002/art.27584 - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

