Characterization of the Neisseria meningitidis Helicase RecG
- PMID: 27736945
- PMCID: PMC5063381
- DOI: 10.1371/journal.pone.0164588
Characterization of the Neisseria meningitidis Helicase RecG
Abstract
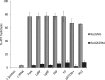
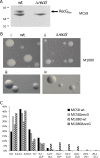
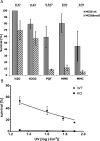
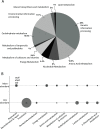
Neisseria meningitidis (Nm) is a Gram-negative oral commensal that opportunistically can cause septicaemia and/or meningitis. Here, we overexpressed, purified and characterized the Nm DNA repair/recombination helicase RecG (RecGNm) and examined its role during genotoxic stress. RecGNm possessed ATP-dependent DNA binding and unwinding activities in vitro on a variety of DNA model substrates including a Holliday junction (HJ). Database searching of the Nm genomes identified 49 single nucleotide polymorphisms (SNPs) in the recGNm including 37 non-synonymous SNPs (nsSNPs), and 7 of the nsSNPs were located in the codons for conserved active site residues of RecGNm. A transient reduction in transformation of DNA was observed in the Nm ΔrecG strain as compared to the wildtype. The gene encoding recGNm also contained an unusually high number of the DNA uptake sequence (DUS) that facilitate transformation in neisserial species. The differentially abundant protein profiles of the Nm wildtype and ΔrecG strains suggest that expression of RecGNm might be linked to expression of other proteins involved in DNA repair, recombination and replication, pilus biogenesis, glycan biosynthesis and ribosomal activity. This might explain the growth defect that was observed in the Nm ΔrecG null mutant.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- Nassif X (2000) Microbiology. A Furtive Pathogen Revealed. Science (80-) 287: 1767–1768. doi: 10.1126/science.287.5459.1767 - DOI - PubMed
-
- Davidsen T, Tønjum T (2006) Meningococcal genome dynamics. Nat Rev Microbiol 4: 11–22. doi: 10.1038/nrmicro1324 - DOI - PubMed
-
- Black CG, Fyfe JA, Davies JK, Black CG, Janet AM, Fyfe JKD (1998) Absence of an SOS-like system in Neisseria gonorrhoeae. Gene 208: 61–66. - PubMed
-
- Nagorska K, Silhan J, Li Y, Pelicic V, Freemont PS, Baldwin GS, et al. (2012) A network of enzymes involved in repair of oxidative DNA damage in Neisseria meningitidis. Mol Microbiol 83: 1064–1079. doi: 10.1111/j.1365-2958.2012.07989.x - DOI - PMC - PubMed
-
- Brosh RM (2013) DNA helicases involved in DNA repair and their roles in cancer. Nat Rev Cancer 13: 542–558. doi: 10.1038/nrc3560 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

