Detecting circular RNAs: bioinformatic and experimental challenges
- PMID: 27739534
- PMCID: PMC5565156
- DOI: 10.1038/nrg.2016.114
Detecting circular RNAs: bioinformatic and experimental challenges
Abstract
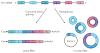
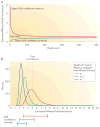
The pervasive expression of circular RNAs (circRNAs) is a recently discovered feature of gene expression in highly diverged eukaryotes. Numerous algorithms that are used to detect genome-wide circRNA expression from RNA sequencing (RNA-seq) data have been developed in the past few years, but there is little overlap in their predictions and no clear gold-standard method to assess the accuracy of these algorithms. We review sources of experimental and bioinformatic biases that complicate the accurate discovery of circRNAs and discuss statistical approaches to address these biases. We conclude with a discussion of the current experimental progress on the topic.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PloS One. 2012;7:e30733. This article provided the first demonstration that circRNA was a ubiquitous and overlooked feature of eukaryotic gene expression. - PMC - PubMed
-
- Zhang XO, et al. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. - PubMed
-
- Szabo L, et al. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015;16:126. The first published circRNA algorithm to develop a statistical score independent of read count for identifying true and false positives. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

