CircRNAs in hematopoiesis and hematological malignancies
- PMID: 27740630
- PMCID: PMC5098259
- DOI: 10.1038/bcj.2016.81
CircRNAs in hematopoiesis and hematological malignancies
Abstract
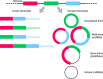
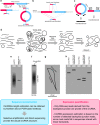
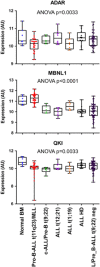
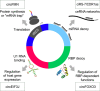
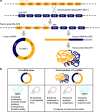
Cell states in hematopoiesis are controlled by master regulators and by complex circuits of a growing family of RNA species impacting cell phenotype maintenance and plasticity. Circular RNAs (circRNAs) are rapidly gaining the status of particularly stable transcriptome members with distinctive qualities. RNA-seq identified thousands of circRNAs with developmental stage- and tissue-specific expression corroborating earlier suggestions that circular isoforms are a natural feature of the cell expression program. CircRNAs are abundantly expressed also in the hematopoietic compartment. There are a number of studies on circRNAs in blood cells, a specific overview is however lacking. In this review we first present current insight in circRNA biogenesis discussing the relevance for hematopoiesis of the highly interleaved processes of splicing and circRNA biogenesis. Regarding molecular functions circRNAs modulate host gene expression, but also compete for binding of microRNAs, RNA-binding proteins or translation initiation and participate in regulatory circuits. We examine circRNA expression in the hematopoietic compartment and in hematologic malignancies and review the recent breakthrough study that identified pathogenic circRNAs derived from leukemia fusion genes. CircRNA high and regulated expression in blood cell types indicate that further studies are warranted to inform the position of these regulators in normal and malignant hematopoiesis.
Figures
References
-
- Li Z, Huang C, Bao C, Chen L, Lin M, Wang X et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015; 22: 256–264. - PubMed
-
- Zhang Y, Zhang X-O, Chen T, Xiang J-F, Yin Q-F, Xing Y-H et al. Circular intronic long noncoding RNAs. Mol Cell 2013; 51: 792–806. - PubMed
-
- Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 2015; 58: 870–885. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

