The Strange, Expanding World of Animal Hepaciviruses
- PMID: 27741408
- PMCID: PMC5523456
- DOI: 10.1146/annurev-virology-100114-055104
The Strange, Expanding World of Animal Hepaciviruses
Abstract
Hepaciviruses and pegiviruses constitute two closely related sister genera of the family Flaviviridae. In the past five years, the known phylogenetic diversity of the hepacivirus genera has absolutely exploded. What was once an isolated infection in humans (and possibly other primates) has now expanded to include horses, rodents, bats, colobus monkeys, cows, and, most recently, catsharks, shedding new light on the genetic diversity and host range of hepaciviruses. Interestingly, despite the identification of these many animal and primate hepaciviruses, the equine hepaciviruses remain the closest genetic relatives of the human hepaciviruses, providing an intriguing clue to the zoonotic source of hepatitis C virus. This review summarizes the significance of these studies and discusses current thinking about the origin and evolution of the animal hepaciviruses as well as their potential usage as surrogate models for the study of hepatitis C virus.
Keywords: animal models; hepatitis C virus; hepegivirus; pegivirus; virome; virus evolution.
Figures

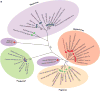


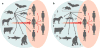
References
-
- Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–62. - PubMed
-
- Simmonds PB, Becher P, Collett MS, Gould EA, Heinz FX, et al. Flaviviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. London: Elsevier; 2012. pp. 1003–20.
-
- Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. 2002;36:S21–29. - PubMed
-
- Yamane D, McGivern DR, Masaki T, Lemon SM. Liver injury and disease pathogenesis in chronic hepatitis C. Curr Top Microbiol Immunol. 2013;369:263–88. - PubMed
-
- Lavanchy D. Evolving epidemiology of hepatitis C virus. Clin Microbiol Infect. 2011;17:107–15. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

