Disentangling the Taxonomy of Rickettsiales and Description of Two Novel Symbionts ("Candidatus Bealeia paramacronuclearis" and "Candidatus Fokinia cryptica") Sharing the Cytoplasm of the Ciliate Protist Paramecium biaurelia
- PMID: 27742680
- PMCID: PMC5118934
- DOI: 10.1128/AEM.02284-16
Disentangling the Taxonomy of Rickettsiales and Description of Two Novel Symbionts ("Candidatus Bealeia paramacronuclearis" and "Candidatus Fokinia cryptica") Sharing the Cytoplasm of the Ciliate Protist Paramecium biaurelia
Abstract
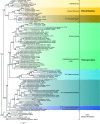
In the past 10 years, the number of endosymbionts described within the bacterial order Rickettsiales has constantly grown. Since 2006, 18 novel Rickettsiales genera inhabiting protists, such as ciliates and amoebae, have been described. In this work, we characterize two novel bacterial endosymbionts from Paramecium collected near Bloomington, IN. Both endosymbiotic species inhabit the cytoplasm of the same host. The Gram-negative bacterium "Candidatus Bealeia paramacronuclearis" occurs in clumps and is frequently associated with the host macronucleus. With its electron-dense cytoplasm and a distinct halo surrounding the cell, it is easily distinguishable from the second smaller symbiont, "Candidatus Fokinia cryptica," whose cytoplasm is electron lucid, lacks a halo, and is always surrounded by a symbiontophorous vacuole. For molecular characterization, the small-subunit rRNA genes were sequenced and used for taxonomic assignment as well as the design of species-specific oligonucleotide probes. Phylogenetic analyses revealed that "Candidatus Bealeia paramacronuclearis" clusters with the so-called "basal" Rickettsiales, and "Candidatus Fokinia cryptica" belongs to "Candidatus Midichloriaceae." We obtained tree topologies showing a separation of Rickettsiales into at least two groups: one represented by the families Rickettsiaceae, Anaplasmataceae, and "Candidatus Midichloriaceae" (RAM clade), and the other represented by "basal Rickettsiales," including "Candidatus Bealeia paramacronuclearis." Therefore, and in accordance with recent publications, we propose to limit the order Rickettsiales to the RAM clade and to raise "basal Rickettsiales" to an independent order, Holosporales ord. nov., inside Alphaproteobacteria, which presently includes four family-level clades. Additionally, we define the family "Candidatus Hepatincolaceae" and redefine the family Holosporaceae IMPORTANCE: In this paper, we provide the characterization of two novel bacterial symbionts inhabiting the same Paramecium host (Ciliophora, Alveolata). Both symbionts belong to "traditional" Rickettsiales, one representing a new species of the genus "Candidatus Fokinia" ("Candidatus Midichloriaceae"), and the other representing a new genus of a "basal" Rickettsiales According to newly characterized sequences and to a critical revision of recent literature, we propose a taxonomic reorganization of "traditional" Rickettsiales that we split into two orders: Rickettsiales sensu stricto and Holosporales ord. nov. This work represents a critical revision, including new records of a group of symbionts frequently occurring in protists and whose biodiversity is still largely underestimated.
Copyright © 2016 Szokoli et al.
Figures





References
-
- Preer LB, Jurand A, Preer JR, Rudman BM. 1972. The classes of kappa in Paramecium aurelia. J Cell Sci 11:581–600. - PubMed
-
- Fujishima M. (ed). 2009. Endosymbionts in Paramecium. Microbiology monographs 12. Springer, Berlin, Germany.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

