Ubiquitous Release of Exosomal Tumor Suppressor miR-6126 from Ovarian Cancer Cells
- PMID: 27742688
- PMCID: PMC5901763
- DOI: 10.1158/0008-5472.CAN-16-0714
Ubiquitous Release of Exosomal Tumor Suppressor miR-6126 from Ovarian Cancer Cells
Erratum in
-
Correction: Ubiquitous Release of Exosomal Tumor Suppressor miR-6126 from Ovarian Cancer Cells.Cancer Res. 2018 Jun 15;78(12):3402. doi: 10.1158/0008-5472.CAN-18-0572. Cancer Res. 2018. PMID: 29907687 No abstract available.
Abstract
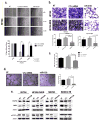
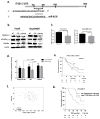
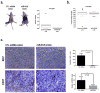
Cancer cells actively promote their tumorigenic behavior by reprogramming gene expression. Loading intraluminal vesicles with specific miRNAs and releasing them into the tumor microenvironment as exosomes is one mechanism of reprogramming whose regulation remains to be elucidated. Here, we report that miR-6126 is ubiquitously released in high abundance from both chemosensitive and chemoresistant ovarian cancer cells via exosomes. Overexpression of miR-6126 was confirmed in healthy ovarian tissue compared with ovarian cancer patient samples and correlated with better overall survival in patients with high-grade serous ovarian cancer. miR-6126 acted as a tumor suppressor by directly targeting integrin-β1, a key regulator of cancer cell metastasis. miR-6126 mimic treatment of cancer cells resulted in increased miR-6126 and decreased integrin-β1 mRNA levels in the exosome. Functional analysis showed that treatment of endothelial cells with miR-6126 mimic significantly reduced tube formation as well as invasion and migration capacities of ovarian cancer cells in vitro Administration of miR-6126 mimic in an orthotopic mouse model of ovarian cancer elicited a relative reduction in tumor growth, proliferating cells, and microvessel density. miR-6126 inhibition promoted oncogenic behavior by leading ovarian cancer cells to release more exosomes. Our findings provide new insights into the role of exosomal miRNA-mediated tumor progression and suggest a new therapeutic approach to disrupt oncogenic phenotypes in tumors. Cancer Res; 76(24); 7194-207. ©2016 AACR.
©2016 American Association for Cancer Research.
Conflict of interest statement
Figures




References
-
- Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J Biol Chem. 1987;262:9412–20. - PubMed
-
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. - PubMed
-
- Abusamra AJ, Zhong Z, Zheng X, Li M, Ichim TE, Chin JL, et al. Tumor exosomes expressing Fas ligand mediate CD8+ T-cell apoptosis. Blood Cells Mol Dis. 2005;35:169–73. - PubMed
-
- Yang L, Wu XH, Wang D, Luo CL, Chen LX. Bladder cancer cell-derived exosomes inhibit tumor cell apoptosis and induce cell proliferation in vitro. Molecular medicine reports. 2013;8:1272–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

