The Brain Proteome of the Ubiquitin Ligase Peli1 Knock-Out Mouse during Experimental Autoimmune Encephalomyelitis
- PMID: 27746629
- PMCID: PMC5061044
- DOI: 10.4172/jpb.1000408
The Brain Proteome of the Ubiquitin Ligase Peli1 Knock-Out Mouse during Experimental Autoimmune Encephalomyelitis
Abstract
The ubiquitin ligase Peli1 has previously been suggested as a potential treatment target in multiple sclerosis. In the multiple sclerosis disease model, experimental autoimmune encephalomyelitis, Peli1 knock-out led to less activated microglia and less inflammation in the central nervous system. Despite being important in microglia, Peli1 expression has also been detected in glial and neuronal cells. In the present study the overall brain proteomes of Peli1 knock-out mice and wild-type mice were compared prior to experimental autoimmune encephalomyelitis induction, at onset of the disease and at disease peak. Brain samples from the frontal hemisphere, peripheral from the extensive inflammatory foci, were analyzed using TMT-labeling of sample pools, and the discovered proteins were verified in individual mice using label-free proteomics. The greatest proteomic differences between Peli1 knock-out and wild-type mice were observed at the disease peak. In Peli1 knock-out a higher degree of antigen presentation, increased activity of adaptive and innate immune cells and alterations to proteins involved in iron metabolism were observed during experimental autoimmune encephalomyelitis. These results unravel global effects to the brain proteome when abrogating Peli1 expression, underlining the importance of Peli1 as a regulator of the immune response also peripheral to inflammatory foci during experimental autoimmune encephalomyelitis. The proteomics data is available in PRIDE with accession PXD003710.
Keywords: Brain; Experimental autoimmune encephalomyelitis (EAE); Label-free proteomics; Multiple sclerosis; Peli1 knock-out; TMT-labeling proteomics.
Figures

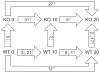

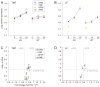
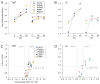


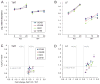
References
-
- Weinshenker BG. Epidemiology of multiple sclerosis. Neurol Clin. 1996;14:291–308. - PubMed
-
- Beeson PB. Age and sex associations of 40 autoimmune diseases. Am J Med. 1994;96:457–462. - PubMed
-
- Harris VK, Sadiq SA. Disease biomarkers in multiple sclerosis: potential for use in therapeutic decision making. Mol Diagn Ther. 2009;13:225–244. - PubMed
-
- Compston A, Coles A. Multiple sclerosis. Lancet. 2008;372:1502–1517. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
