Comparative Genomic Analysis among Four Representative Isolates of Phytophthora sojae Reveals Genes under Evolutionary Selection
- PMID: 27746768
- PMCID: PMC5042962
- DOI: 10.3389/fmicb.2016.01547
Comparative Genomic Analysis among Four Representative Isolates of Phytophthora sojae Reveals Genes under Evolutionary Selection
Abstract
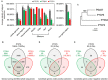
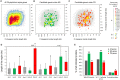
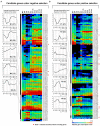
Comparative genomic analysis is useful for identifying genes affected by evolutionary selection and for studying adaptive variation in gene functions. In Phytophthora sojae, a model oomycete plant pathogen, the related study is lacking. We compared sequence data among four isolates of P. sojae, which represent its four major genotypes. These isolates exhibited >99.688%, >99.864%, and >98.981% sequence identities at genome, gene, and non-gene regions, respectively. One hundred and fifty-three positive selection and 139 negative selection candidate genes were identified. Between the two categories of genes, the positive selection genes were flanked by larger intergenic regions, poorly annotated in function, and less conserved; they had relatively lower transcription levels but many genes had increased transcripts during infection. Genes coding for predicted secreted proteins, particularly effectors, were overrepresented in positive selection. Several RxLR effector genes were identified as positive selection genes, exhibiting much stronger positive selection levels. In addition, candidate genes with presence/absence polymorphism were analyzed. This study provides a landscape of genomic variation among four representative P. sojae isolates and characterized several evolutionary selection-affected gene candidates. The results suggest a relatively covert two-speed genome evolution pattern in P. sojae and will provide clues for identification of new virulence factors in the oomycete plant pathogens.
Keywords: Phytophthora; comparative genomic analysis; effectors; evolution; positive selection.
Figures
Similar articles
-
Genome-wide identification of long non-coding RNAs suggests a potential association with effector gene transcription in Phytophthora sojae.Mol Plant Pathol. 2018 Sep;19(9):2177-2186. doi: 10.1111/mpp.12692. Epub 2018 Jul 17. Mol Plant Pathol. 2018. PMID: 29665235 Free PMC article.
-
Whole Genome Re-sequencing Reveals Natural Variation and Adaptive Evolution of Phytophthora sojae.Front Microbiol. 2019 Nov 29;10:2792. doi: 10.3389/fmicb.2019.02792. eCollection 2019. Front Microbiol. 2019. PMID: 31849921 Free PMC article.
-
Copy number variation and transcriptional polymorphisms of Phytophthora sojae RXLR effector genes Avr1a and Avr3a.PLoS One. 2009;4(4):e5066. doi: 10.1371/journal.pone.0005066. Epub 2009 Apr 3. PLoS One. 2009. PMID: 19343173 Free PMC article.
-
Phenotypic diversification by gene silencing in Phytophthora plant pathogens.Commun Integr Biol. 2013 Nov 1;6(6):e25890. doi: 10.4161/cib.25890. Epub 2013 Jul 26. Commun Integr Biol. 2013. PMID: 24563702 Free PMC article. Review.
-
Phytophthora sojae effectors orchestrate warfare with host immunity.Curr Opin Microbiol. 2018 Dec;46:7-13. doi: 10.1016/j.mib.2018.01.008. Epub 2018 Feb 22. Curr Opin Microbiol. 2018. PMID: 29454192 Review.
Cited by
-
Comparative Genomic Analysis Reveals Genetic Variation and Adaptive Evolution in the Pathogenicity-Related Genes of Phytophthora capsici.Front Microbiol. 2021 Aug 13;12:694136. doi: 10.3389/fmicb.2021.694136. eCollection 2021. Front Microbiol. 2021. PMID: 34484141 Free PMC article.
-
Genome-wide identification of long non-coding RNAs suggests a potential association with effector gene transcription in Phytophthora sojae.Mol Plant Pathol. 2018 Sep;19(9):2177-2186. doi: 10.1111/mpp.12692. Epub 2018 Jul 17. Mol Plant Pathol. 2018. PMID: 29665235 Free PMC article.
-
Whole Genome Re-sequencing Reveals Natural Variation and Adaptive Evolution of Phytophthora sojae.Front Microbiol. 2019 Nov 29;10:2792. doi: 10.3389/fmicb.2019.02792. eCollection 2019. Front Microbiol. 2019. PMID: 31849921 Free PMC article.
-
Comparative Genomics and Gene Pool Analysis Reveal the Decrease of Genome Diversity and Gene Number in Rice Blast Fungi by Stable Adaption with Rice.J Fungi (Basel). 2021 Dec 22;8(1):5. doi: 10.3390/jof8010005. J Fungi (Basel). 2021. PMID: 35049945 Free PMC article.
-
Molecular mechanisms of Phytophthora sojae avirulence effectors escaping host recognition.Front Microbiol. 2023 Jan 9;13:1111774. doi: 10.3389/fmicb.2022.1111774. eCollection 2022. Front Microbiol. 2023. PMID: 36699593 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

