Genome scans for divergent selection in natural populations of the widespread hardwood species Eucalyptus grandis (Myrtaceae) using microsatellites
- PMID: 27748400
- PMCID: PMC5066178
- DOI: 10.1038/srep34941
Genome scans for divergent selection in natural populations of the widespread hardwood species Eucalyptus grandis (Myrtaceae) using microsatellites
Abstract
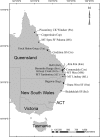
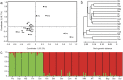
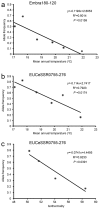
Identification of loci or genes under natural selection is important for both understanding the genetic basis of local adaptation and practical applications, and genome scans provide a powerful means for such identification purposes. In this study, genome-wide simple sequence repeats markers (SSRs) were used to scan for molecular footprints of divergent selection in Eucalyptus grandis, a hardwood species occurring widely in costal areas from 32° S to 16° S in Australia. High population diversity levels and weak population structure were detected with putatively neutral genomic SSRs. Using three FST outlier detection methods, a total of 58 outlying SSRs were collectively identified as loci under divergent selection against three non-correlated climatic variables, namely, mean annual temperature, isothermality and annual precipitation. Using a spatial analysis method, nine significant associations were revealed between FST outlier allele frequencies and climatic variables, involving seven alleles from five SSR loci. Of the five significant SSRs, two (EUCeSSR1044 and Embra394) contained alleles of putative genes with known functional importance for response to climatic factors. Our study presents critical information on the population diversity and structure of the important woody species E. grandis and provides insight into the adaptive responses of perennial trees to climatic variations.
Figures
Similar articles
-
Signatures of diversifying selection at EST-SSR loci and association with climate in natural Eucalyptus populations.Mol Ecol. 2013 Oct;22(20):5112-29. doi: 10.1111/mec.12463. Mol Ecol. 2013. PMID: 24118117
-
Evidence of genomic adaptation to climate in Eucalyptus microcarpa: Implications for adaptive potential to projected climate change.Mol Ecol. 2017 Nov;26(21):6002-6020. doi: 10.1111/mec.14341. Epub 2017 Oct 7. Mol Ecol. 2017. PMID: 28862778
-
Genome-wide scans detect adaptation to aridity in a widespread forest tree species.Mol Ecol. 2014 May;23(10):2500-13. doi: 10.1111/mec.12751. Mol Ecol. 2014. PMID: 24750317
-
Eucalyptus applied genomics: from gene sequences to breeding tools.New Phytol. 2008;179(4):911-929. doi: 10.1111/j.1469-8137.2008.02503.x. Epub 2008 Jun 5. New Phytol. 2008. PMID: 18537893 Review.
-
Microsatellite resources of Eucalyptus: current status and future perspectives.Bot Stud. 2014 Dec;55(1):73. doi: 10.1186/s40529-014-0073-3. Epub 2014 Oct 25. Bot Stud. 2014. PMID: 28510953 Free PMC article. Review.
Cited by
-
Seascape Genomics of the Sugar Kelp Saccharina latissima along the North Eastern Atlantic Latitudinal Gradient.Genes (Basel). 2020 Dec 13;11(12):1503. doi: 10.3390/genes11121503. Genes (Basel). 2020. PMID: 33322137 Free PMC article.
-
Thirteen years under arid conditions: exploring marker-trait associations in Eucalyptus cladocalyx for complex traits related to flowering, stem form and growth.Breed Sci. 2018 Jun;68(3):367-374. doi: 10.1270/jsbbs.17131. Epub 2018 Jun 29. Breed Sci. 2018. PMID: 30100804 Free PMC article.
-
Genetic relationship analysis and core collection construction of Eucalyptus grandis from Dongmen improved variety base: the largest eucalypt germplasm resource in China.BMC Plant Biol. 2024 Dec 23;24(1):1240. doi: 10.1186/s12870-024-05970-0. BMC Plant Biol. 2024. PMID: 39716061 Free PMC article.
-
Testing for fitness epistasis in a transplant experiment identifies a candidate adaptive locus in Timema stick insects.Philos Trans R Soc Lond B Biol Sci. 2022 Jul 18;377(1855):20200508. doi: 10.1098/rstb.2020.0508. Epub 2022 May 30. Philos Trans R Soc Lond B Biol Sci. 2022. PMID: 35634927 Free PMC article.
-
The first identification of genomic loci in plants associated with resistance to galling insects: a case study in Eucalyptus L'Hér. (Myrtaceae).Sci Rep. 2018 Feb 2;8(1):2319. doi: 10.1038/s41598-018-20780-9. Sci Rep. 2018. PMID: 29396525 Free PMC article.
References
-
- Davis M. B. & Shaw R. G. Range shifts and adaptive responses to Quaternary climate change. Science 292, 673–679 (2001). - PubMed
-
- Jump A. S. & Peñuelas J. Running to stand still: adaptation and the response of plants to rapid climate change. Ecol. Lett. 8, 1010–1020 (2005). - PubMed
-
- Aitken S. N. & Whitlock M. C. Assisted gene flow to facilitate local adaptation to climate change. Ann. Rev. Ecol. Evol. Syst. 44, 367–388 (2013).
-
- Jackson S. T. & Overpeck J. T. Responses of plant populations and communities to environmental changes of the late Quaternary. Paleobiology 26, 194–220 (2000).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

