MicroRNA-127 targeting of mitoNEET inhibits neurite outgrowth, induces cell apoptosis and contributes to physiological dysfunction after spinal cord transection
- PMID: 27748416
- PMCID: PMC5066253
- DOI: 10.1038/srep35205
MicroRNA-127 targeting of mitoNEET inhibits neurite outgrowth, induces cell apoptosis and contributes to physiological dysfunction after spinal cord transection
Erratum in
-
Erratum: MicroRNA-127 targeting of mitoNEET inhibits neurite outgrowth, induces cell apoptosis and contributes to physiological dysfunction after spinal cord transection.Sci Rep. 2017 Jan 19;7:39570. doi: 10.1038/srep39570. Sci Rep. 2017. PMID: 28102200 Free PMC article. No abstract available.
Abstract
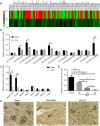
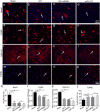
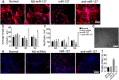
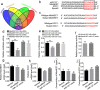
Neuroregeneration and apoptosis are two important pathophysiologic changes after spinal cord injury (SCI), but their underlying mechanisms remain unclear. MicroRNAs (miRNAs) play a crucial role in the regulation of neuroregeneration and neuronal apoptosis, research areas that have been greatly expanded in recent years. Here, using miRNA arrays to profile miRNA transcriptomes, we demonstrated that miR-127-3p was significantly down-regulated after spinal cord transection (SCT). Then, bioinformatics analyses and experimental detection showed that miR-127-3p exhibited specific effects on the regulation of neurite outgrowth and the induction of neuronal apoptosis by regulating the expression of the mitochondrial membrane protein mitoNEET. Moreover, knockdown of MitoNEET leaded to neuronal loss and apoptosis in primary cultured spinal neurons. This study therefore revealed that miR-127-3p, which targets mitoNEET, plays a vital role in regulating neurite outgrowth and neuronal apoptosis after SCT. Thus, modificatioin of the mitoNEET expression, such as mitoNEET activition may provide a new strategy for the treatment of SCI in preclinical trials.
Figures


References
-
- Tederko P., Krasuski M., Kiwerski J., Nyka I. & Bialoszewski D. Strategies for neuroprotection following spinal cord injury.Ortopedia, traumatologia, rehabilitacja. 11, 103–110 (2009). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

