Binding of cellulose binding modules reveal differences between cellulose substrates
- PMID: 27748440
- PMCID: PMC5066208
- DOI: 10.1038/srep35358
Binding of cellulose binding modules reveal differences between cellulose substrates
Abstract
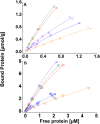
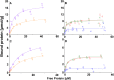
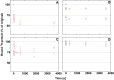
The interaction between cellulase enzymes and their substrates is of central importance to several technological and scientific challenges. Here we report that the binding of cellulose binding modules (CBM) from Trichoderma reesei cellulases Cel6A and Cel7A show a major difference in how they interact with substrates originating from wood compared to bacterial cellulose. We found that the CBM from TrCel7A recognizes the two substrates differently and as a consequence shows an unexpected way of binding. We show that the substrate has a large impact on the exchange rate of the studied CBM, and moreover, CBM-TrCel7A seems to have an additional mode of binding on wood derived cellulose but not on cellulose originating from bacterial source. This mode is not seen in double CBM (DCBM) constructs comprising both CBM-TrCel7A and CBM-TrCel6A. The linker length of DCBMs affects the binding properties, and slows down the exchange rates of the proteins and thus, can be used to analyze the differences between the single CBM. These results have impact on the cellulase research and offer new understanding on how these industrially relevant enzymes act.
Figures
Similar articles
-
Single-molecule Imaging Analysis of Binding, Processive Movement, and Dissociation of Cellobiohydrolase Trichoderma reesei Cel6A and Its Domains on Crystalline Cellulose.J Biol Chem. 2016 Oct 21;291(43):22404-22413. doi: 10.1074/jbc.M116.752048. Epub 2016 Sep 8. J Biol Chem. 2016. PMID: 27609516 Free PMC article.
-
Inter-domain Synergism Is Required for Efficient Feeding of Cellulose Chain into Active Site of Cellobiohydrolase Cel7A.J Biol Chem. 2016 Dec 9;291(50):26013-26023. doi: 10.1074/jbc.M116.756007. Epub 2016 Oct 25. J Biol Chem. 2016. PMID: 27780868 Free PMC article.
-
The tryptophan residue at the active site tunnel entrance of Trichoderma reesei cellobiohydrolase Cel7A is important for initiation of degradation of crystalline cellulose.J Biol Chem. 2013 May 10;288(19):13503-10. doi: 10.1074/jbc.M113.452623. Epub 2013 Mar 26. J Biol Chem. 2013. PMID: 23532843 Free PMC article.
-
Glycosylated linkers in multimodular lignocellulose-degrading enzymes dynamically bind to cellulose.Proc Natl Acad Sci U S A. 2013 Sep 3;110(36):14646-51. doi: 10.1073/pnas.1309106110. Epub 2013 Aug 19. Proc Natl Acad Sci U S A. 2013. PMID: 23959893 Free PMC article.
-
Functional analysis of chimeric TrCel6A enzymes with different carbohydrate binding modules.Protein Eng Des Sel. 2019 Dec 31;32(9):401-409. doi: 10.1093/protein/gzaa003. Protein Eng Des Sel. 2019. PMID: 32100026
Cited by
-
Binding Forces of Cellulose Binding Modules on Cellulosic Nanomaterials.Biomacromolecules. 2019 Feb 11;20(2):769-777. doi: 10.1021/acs.biomac.8b01346. Epub 2019 Jan 31. Biomacromolecules. 2019. PMID: 30657665 Free PMC article.
-
A novel accessory protein ArCel5 from cellulose-gelatinizing fungus Arthrobotrys sp. CX1.Bioresour Bioprocess. 2022 Mar 21;9(1):27. doi: 10.1186/s40643-022-00519-1. Bioresour Bioprocess. 2022. PMID: 38647580 Free PMC article.
-
Fungal-type carbohydrate binding modules from the coccolithophore Emiliania huxleyi show binding affinity to cellulose and chitin.PLoS One. 2018 May 21;13(5):e0197875. doi: 10.1371/journal.pone.0197875. eCollection 2018. PLoS One. 2018. PMID: 29782536 Free PMC article.
-
Bioengineered textiles with peptide binders that capture SARS-CoV-2 viral particles.Commun Mater. 2022;3(1):54. doi: 10.1038/s43246-022-00278-8. Epub 2022 Aug 15. Commun Mater. 2022. PMID: 35991518 Free PMC article.
-
In Vitro Evaluation of Enriched Brewers' Spent Grains Using Bacillus subtilis WX-17 as Potential Functional Food Ingredients.Appl Biochem Biotechnol. 2021 Feb;193(2):349-362. doi: 10.1007/s12010-020-03424-5. Epub 2020 Sep 24. Appl Biochem Biotechnol. 2021. PMID: 32968964
References
-
- Henrissat B. Cellulases and their interaction with cellulose. Cellulose 1, 169–196 (1994).
-
- Várnai A. et al. Carbohydrate-Binding Modules of Fungal Cellulases. Occurrence in Nature, Function, and Relevance in Industrial Biomass Conversion. Adv. Appl. Microbiol. 88, 103–165 (2014). - PubMed
-
- Hägglund P. et al. A cellulose-binding module of the Trichoderma reesei β-mannanase Man5A increases the mannan-hydrolysis of complex substrates. J. Biotechnol. 101, 37–48 (2003). - PubMed
-
- Raghothama S. et al. Solution structure of the CBM10 cellulose binding module from pseudomonas xylanase A. Biochemistry 39, 978–984 (2000). - PubMed
-
- Stern D. L. The genetic causes of convergent evolution. Nat. Rev. Genet. 14, 751–764 (2013). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

