Antagonistic negative and positive neurons of the basolateral amygdala
- PMID: 27749826
- PMCID: PMC5493320
- DOI: 10.1038/nn.4414
Antagonistic negative and positive neurons of the basolateral amygdala
Abstract
The basolateral amygdala (BLA) is a site of convergence of negative and positive stimuli and is critical for emotional behaviors and associations. However, the neural substrate for negative and positive behaviors and relationship between negative and positive representations in the basolateral amygdala are unknown. Here we identify two genetically distinct, spatially segregated populations of excitatory neurons in the mouse BLA that participate in valence-specific behaviors and are connected through mutual inhibition. These results identify a genetically defined neural circuit for the antagonistic control of emotional behaviors and memories.
Figures
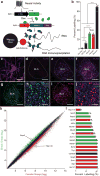

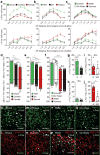
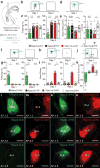
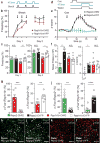


References
-
- Pitkanen A, Savander V, LeDoux JE. Organization of intra-amygdaloid circuitries in the rat: an emerging framework for understanding functions of the amygdala. Trends in neurosciences. 1997;20:517–523. - PubMed
-
- McDonald AJ. Neuronal organization of the lateral and basolateral amygdaloid nuclei in the rat. The Journal of comparative neurology. 1984;222:589–606. - PubMed
-
- Swanson LW, Petrovich GD. What is the amygdala? Trends in neurosciences. 1998;21:323–331. - PubMed
-
- Hall E. The amygdala of the cat: a Golgi study. Zeitschrift fur Zellforschung und mikroskopische Anatomie. 1972;134:439–458. - PubMed
-
- McDonald AJ. Neurons of the lateral and basolateral amygdaloid nuclei: a Golgi study in the rat. The Journal of comparative neurology. 1982;212:293–312. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

