Spatial intratumoral heterogeneity and temporal clonal evolution in esophageal squamous cell carcinoma
- PMID: 27749841
- PMCID: PMC5127772
- DOI: 10.1038/ng.3683
Spatial intratumoral heterogeneity and temporal clonal evolution in esophageal squamous cell carcinoma
Abstract
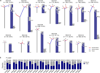
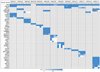
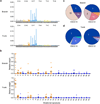
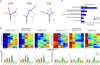
Esophageal squamous cell carcinoma (ESCC) is among the most common malignancies, but little is known about its spatial intratumoral heterogeneity (ITH) and temporal clonal evolutionary processes. To address this, we performed multiregion whole-exome sequencing on 51 tumor regions from 13 ESCC cases and multiregion global methylation profiling for 3 of these 13 cases. We found an average of 35.8% heterogeneous somatic mutations with strong evidence of ITH. Half of the driver mutations located on the branches of tumor phylogenetic trees targeted oncogenes, including PIK3CA, NFE2L2 and MTOR, among others. By contrast, the majority of truncal and clonal driver mutations occurred in tumor-suppressor genes, including TP53, KMT2D and ZNF750, among others. Interestingly, phyloepigenetic trees robustly recapitulated the topological structures of the phylogenetic trees, indicating a possible relationship between genetic and epigenetic alterations. Our integrated investigations of spatial ITH and clonal evolution provide an important molecular foundation for enhanced understanding of tumorigenesis and progression in ESCC.
Figures
Comment in
-
Intratumoral heterogeneity of esophageal squamous cell carcinoma.J Thorac Dis. 2017 May;9(5):E465-E467. doi: 10.21037/jtd.2017.04.54. J Thorac Dis. 2017. PMID: 28616310 Free PMC article. No abstract available.
References
-
- Torre LA, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. - PubMed
-
- Ferlay J, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. - PubMed
-
- Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2252. - PubMed
-
- Song Y, et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509:91–95. - PubMed
METHODS-ONLY REFERENCES
-
- Chen W, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

