Morphological and Transcriptomic Analysis of a Beetle Chemosensory System Reveals a Gnathal Olfactory Center
- PMID: 27751175
- PMCID: PMC5067906
- DOI: 10.1186/s12915-016-0304-z
Morphological and Transcriptomic Analysis of a Beetle Chemosensory System Reveals a Gnathal Olfactory Center
Abstract
Background: The red flour beetle Tribolium castaneum is an emerging insect model organism representing the largest insect order, Coleoptera, which encompasses several serious agricultural and forest pests. Despite the ecological and economic importance of beetles, most insect olfaction studies have so far focused on dipteran, lepidopteran, or hymenopteran systems.
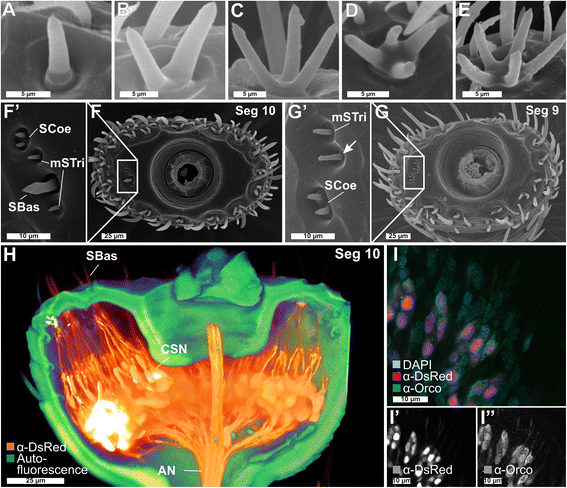
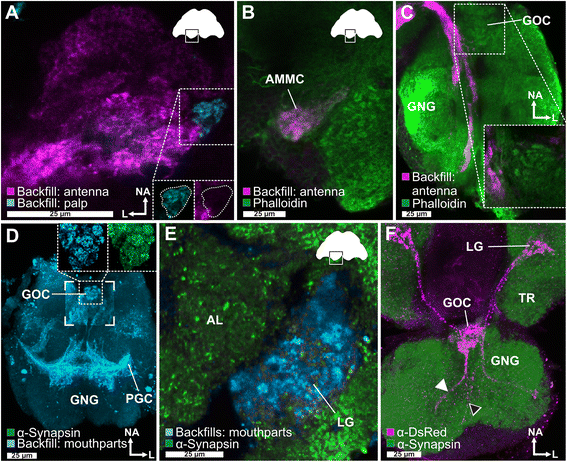
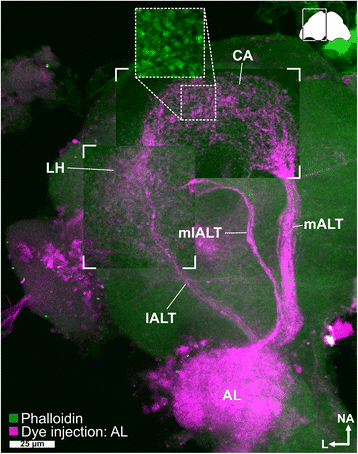
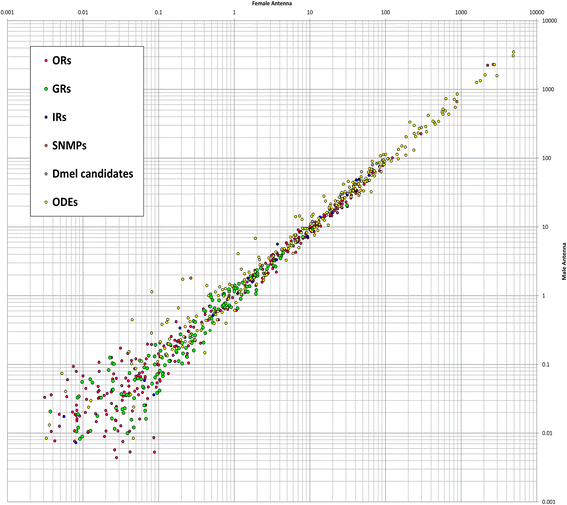
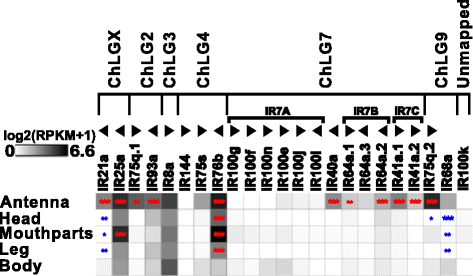
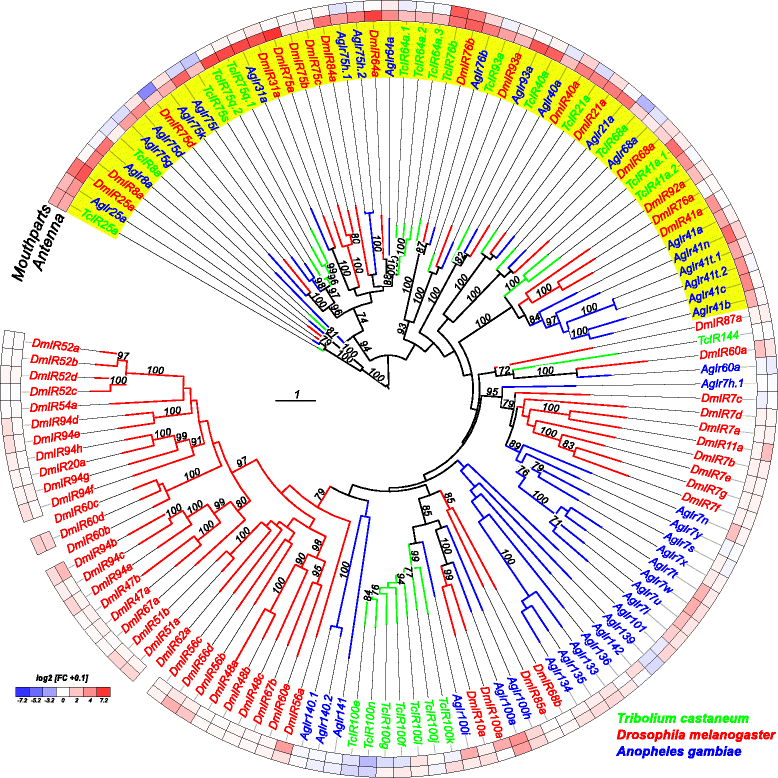
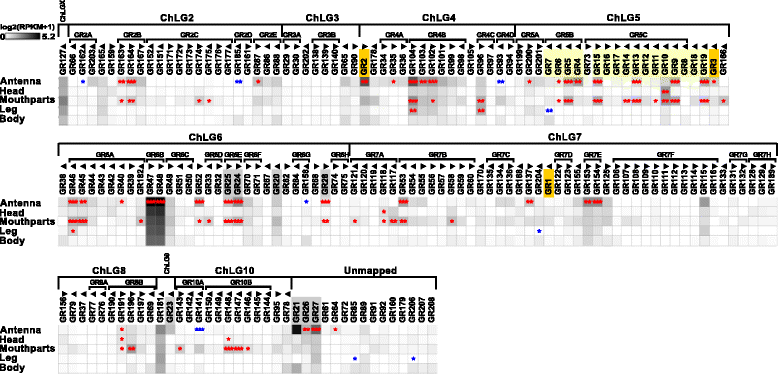
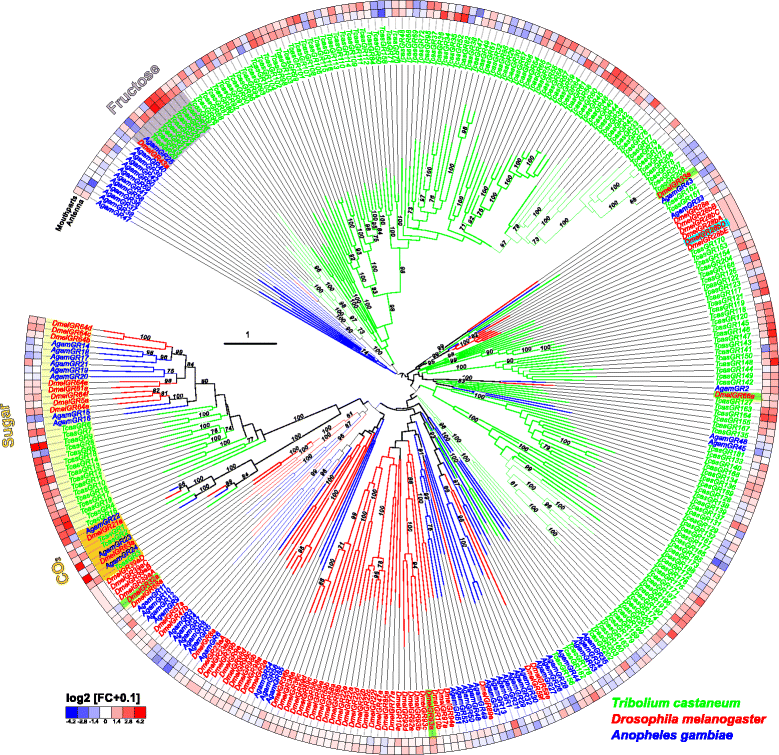
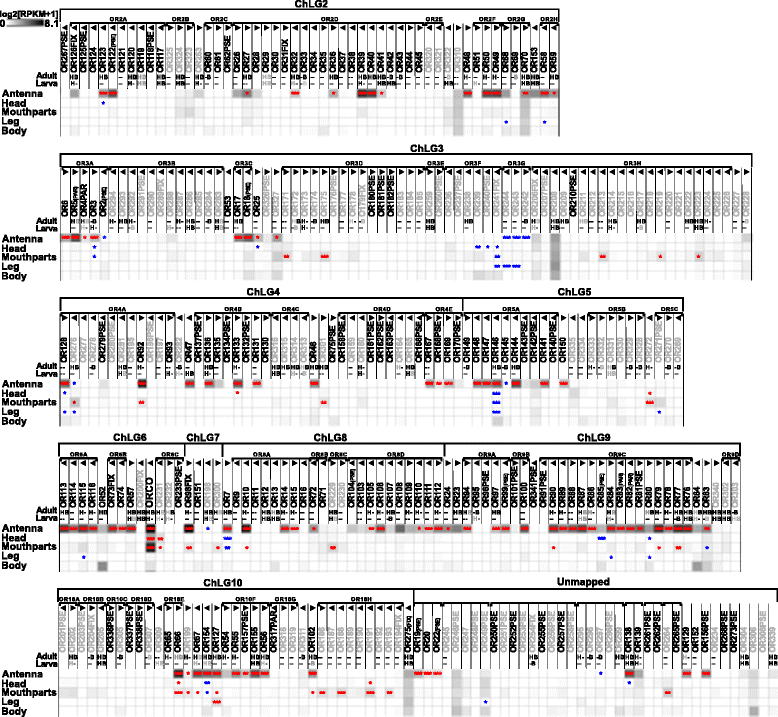
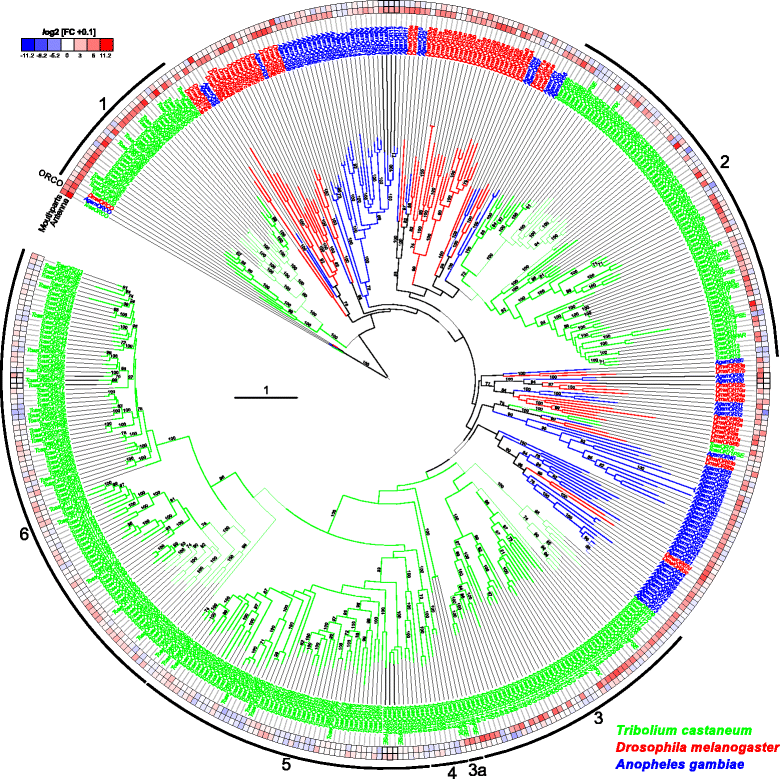
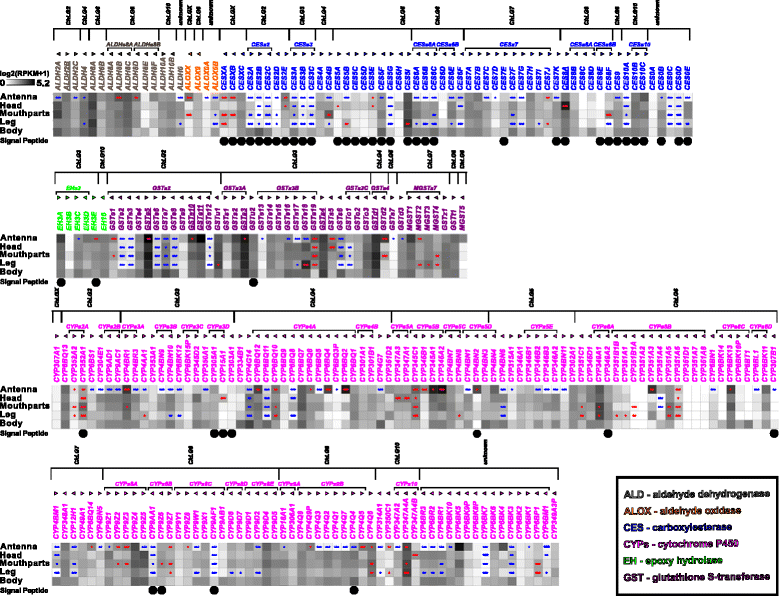
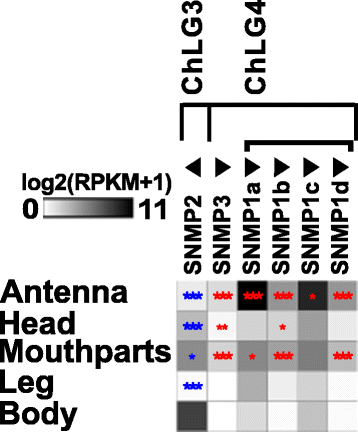
Results: Here, we present the first detailed morphological description of a coleopteran olfactory pathway in combination with genome-wide expression analysis of the relevant gene families involved in chemoreception. Our study revealed that besides the antennae, also the mouthparts are highly involved in olfaction and that their respective contribution is processed separately. In this beetle, olfactory sensory neurons from the mouthparts project to the lobus glomerulatus, a structure so far only characterized in hemimetabolous insects, as well as to a so far non-described unpaired glomerularly organized olfactory neuropil in the gnathal ganglion, which we term the gnathal olfactory center. The high number of functional odorant receptor genes expressed in the mouthparts also supports the importance of the maxillary and labial palps in olfaction of this beetle. Moreover, gustatory perception seems equally distributed between antenna and mouthparts, since the number of expressed gustatory receptors is similar for both organs.
Conclusions: Our analysis of the T. castaneum chemosensory system confirms that olfactory and gustatory perception are not organotopically separated to the antennae and mouthparts, respectively. The identification of additional olfactory processing centers, the lobus glomerulatus and the gnathal olfactory center, is in contrast to the current picture that in holometabolous insects all olfactory inputs allegedly converge in the antennal lobe. These findings indicate that Holometabola have evolved a wider variety of solutions to chemoreception than previously assumed.
Keywords: Tribolium castaneum; chemoreception; gustation; insect; lobus glomerulatus; neuroanatomy; olfaction.
Figures




Similar articles
-
Anatomic and neurochemical analysis of the palpal olfactory system in the red flour beetle Tribolium castaneum, HERBST.Front Cell Neurosci. 2023 Feb 23;17:1097462. doi: 10.3389/fncel.2023.1097462. eCollection 2023. Front Cell Neurosci. 2023. PMID: 36998268 Free PMC article.
-
Tissue-specific transcriptomics, chromosomal localization, and phylogeny of chemosensory and odorant binding proteins from the red flour beetle Tribolium castaneum reveal subgroup specificities for olfaction or more general functions.BMC Genomics. 2014 Dec 18;15:1141. doi: 10.1186/1471-2164-15-1141. BMC Genomics. 2014. PMID: 25523483 Free PMC article.
-
Antennal transcriptome analysis of the chemosensory gene families in the tree killing bark beetles, Ips typographus and Dendroctonus ponderosae (Coleoptera: Curculionidae: Scolytinae).BMC Genomics. 2013 Mar 21;14:198. doi: 10.1186/1471-2164-14-198. BMC Genomics. 2013. PMID: 23517120 Free PMC article.
-
Molecular evolution of the major chemosensory gene families in insects.Heredity (Edinb). 2009 Sep;103(3):208-16. doi: 10.1038/hdy.2009.55. Epub 2009 May 13. Heredity (Edinb). 2009. PMID: 19436326 Review.
-
Elements of olfactory reception in adult Drosophila melanogaster.Anat Rec (Hoboken). 2013 Sep;296(9):1477-88. doi: 10.1002/ar.22747. Epub 2013 Jul 31. Anat Rec (Hoboken). 2013. PMID: 23904114 Review.
Cited by
-
Adult neurogenesis in the mushroom bodies of red flour beetles (Tribolium castaneum, HERBST) is influenced by the olfactory environment.Sci Rep. 2020 Jan 23;10(1):1090. doi: 10.1038/s41598-020-57639-x. Sci Rep. 2020. PMID: 31974446 Free PMC article.
-
Odor-Induced Vomiting Is Combinatorially Triggered by Palp Olfactory Receptor Neurons That Project to the Lobus Glomerulatus in Locust Brain.Front Physiol. 2022 Apr 20;13:855522. doi: 10.3389/fphys.2022.855522. eCollection 2022. Front Physiol. 2022. PMID: 35514359 Free PMC article.
-
The Two Main Olfactory Receptor Families in Drosophila, ORs and IRs: A Comparative Approach.Front Cell Neurosci. 2018 Aug 30;12:253. doi: 10.3389/fncel.2018.00253. eCollection 2018. Front Cell Neurosci. 2018. PMID: 30214396 Free PMC article. Review.
-
Expanding the genetic toolkit of Tribolium castaneum.PLoS One. 2018 Apr 12;13(4):e0195977. doi: 10.1371/journal.pone.0195977. eCollection 2018. PLoS One. 2018. PMID: 29649291 Free PMC article.
-
Mutually Exclusive Expression of Closely Related Odorant-Binding Proteins 9A and 9B in the Antenna of the Red Flour Beetle Tribolium castaneum.Biomolecules. 2021 Oct 12;11(10):1502. doi: 10.3390/biom11101502. Biomolecules. 2021. PMID: 34680135 Free PMC article.
References
-
- Visser JH. Host odor perception in phytophagous insects. Annu Rev Entomol. 1986;31:121–44. doi: 10.1146/annurev.en.31.010186.001005. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

