A natural barrier to lateral gene transfer from prokaryotes to eukaryotes revealed from genomes: the 70 % rule
- PMID: 27751184
- PMCID: PMC5067920
- DOI: 10.1186/s12915-016-0315-9
A natural barrier to lateral gene transfer from prokaryotes to eukaryotes revealed from genomes: the 70 % rule
Abstract
Background: The literature harbors many claims for lateral gene transfer (LGT) from prokaryotes to eukaryotes. Such claims are typically founded in analyses of genome sequences. It is undisputed that many genes entered the eukaryotic lineage via the origin of mitochondria and the origin of plastids. Claims for lineage-specific LGT to eukaryotes outside the context of organelle origins and claims of continuous LGT to eukaryotic lineages are more problematic. If eukaryotes acquire genes from prokaryotes continuously during evolution, then sequenced eukaryote genomes should harbor evidence for recent LGT, like prokaryotic genomes do.
Results: Here we devise an approach to investigate 30,358 eukaryotic sequences in the context of 1,035,375 prokaryotic homologs among 2585 phylogenetic trees containing homologs from prokaryotes and eukaryotes. Prokaryote genomes reflect a continuous process of gene acquisition and inheritance, with abundant recent acquisitions showing 80-100 % amino acid sequence identity to their phylogenetic sister-group homologs from other phyla. By contrast, eukaryote genomes show no evidence for either continuous or recent gene acquisitions from prokaryotes. We find that, in general, genes in eukaryotic genomes that share ≥70 % amino acid identity to prokaryotic homologs are genome-specific; that is, they are not found outside individual genome assemblies.
Conclusions: Our analyses indicate that eukaryotes do not acquire genes through continual LGT like prokaryotes do. We propose a 70 % rule: Coding sequences in eukaryotic genomes that share more than 70 % amino acid sequence identity to prokaryotic homologs are most likely assembly or annotation artifacts. The findings further uncover that the role of differential loss in eukaryote genome evolution has been vastly underestimated.
Figures
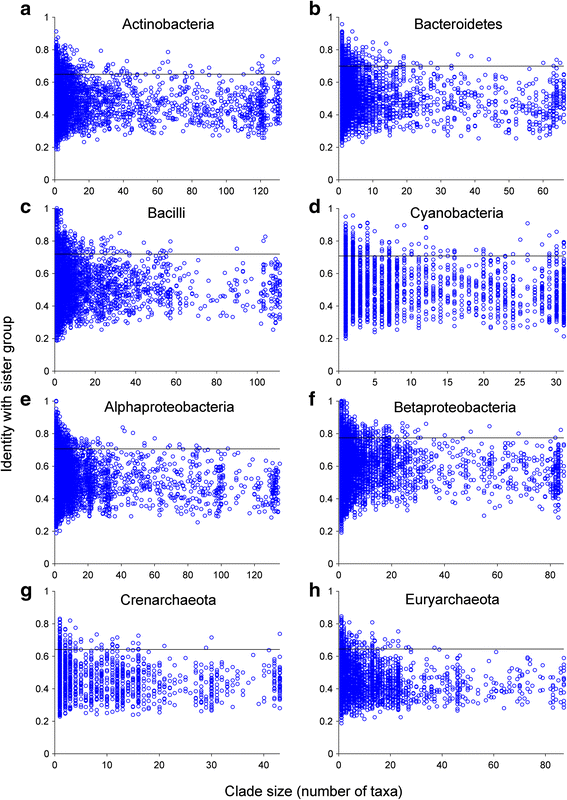
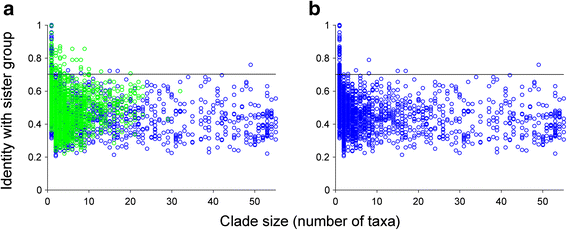

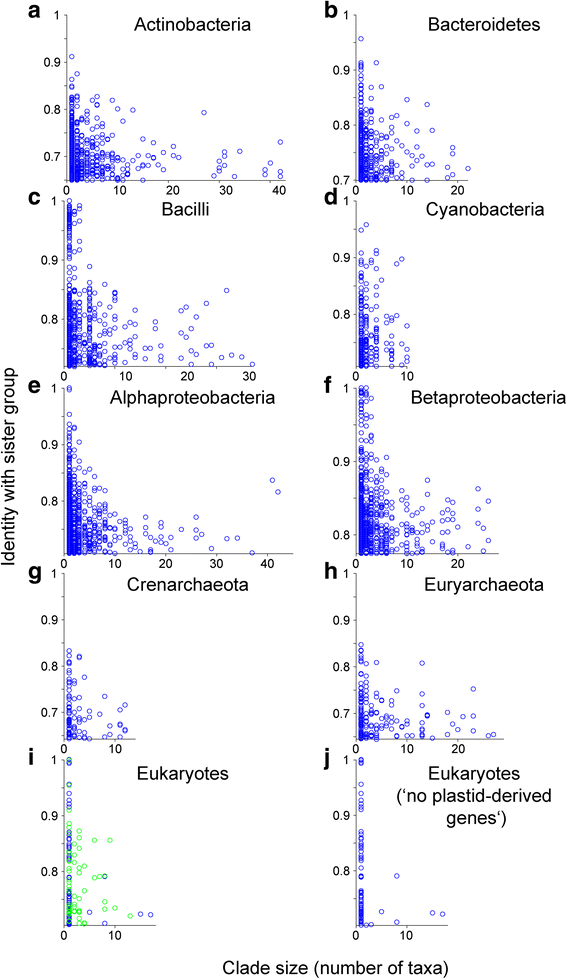

Comment in
-
Lateral gene transfer in eukaryotes: tip of the iceberg or of the ice cube?BMC Biol. 2016 Nov 18;14(1):101. doi: 10.1186/s12915-016-0330-x. BMC Biol. 2016. PMID: 27863503 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

