Structural Dynamics in Ras and Related Proteins upon Nucleotide Switching
- PMID: 27751724
- PMCID: PMC5661953
- DOI: 10.1016/j.jmb.2016.10.017
Structural Dynamics in Ras and Related Proteins upon Nucleotide Switching
Abstract
Structural dynamics of Ras proteins contributes to their activity in signal transduction cascades. Directly targeting Ras proteins with small molecules may rely on the movement of a conserved structural motif, switch II. To understand Ras signaling and advance Ras-targeting strategies, experimental methods to measure Ras dynamics are required. Here, we demonstrate the utility of hydrogen-deuterium exchange (HDX) mass spectrometry (MS) to measure Ras dynamics by studying representatives from two branches of the Ras superfamily, Ras and Rho. A comparison of differential deuterium exchange between active (GMPPNP-bound) and inactive (GDP-bound) proteins revealed differences between the families, with the most notable differences occurring in the phosphate-binding loop and switch II. The P-loop exchange signature correlated with switch II dynamics observed in molecular dynamics simulations focused on measuring main-chain movement. HDX provides a means of evaluating Ras protein dynamics, which may be useful for understanding the mechanisms of Ras signaling, including activated signaling of pathologic mutants, and for targeting strategies that rely on protein dynamics.
Keywords: Rho; guanosine diphosphate (GDP); hydrogen–deuterium exchange mass spectrometry; protein dynamics; signal transduction.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures

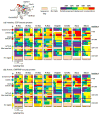

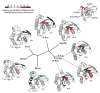
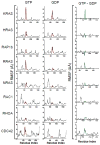
References
-
- Laufs U, Liao JK. Targeting Rho in cardiovascular disease. Circ Res. 2000;87:526–8. - PubMed
-
- Seabra MC, Mules EH, Hume AN. Rab GTPases, intracellular traffic and disease. Trends Mol Med. 2002;8:23–30. - PubMed
-
- Linseman DA, Loucks FA. Diverse roles of Rho family GTPases in neuronal development, survival, and death. Front Biosci. 2007;13:657–76. - PubMed
-
- Bos JL. Ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–9. - PubMed
-
- Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

