Low-mass-ion discriminant equation (LOME) for ovarian cancer screening
- PMID: 27752286
- PMCID: PMC5059959
- DOI: 10.1186/s13040-016-0111-7
Low-mass-ion discriminant equation (LOME) for ovarian cancer screening
Abstract
Background: A low-mass-ion discriminant equation (LOME) was constructed to investigate whether systematic low-mass-ion (LMI) profiling could be applied to ovarian cancer (OVC) screening.
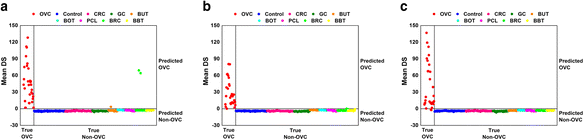
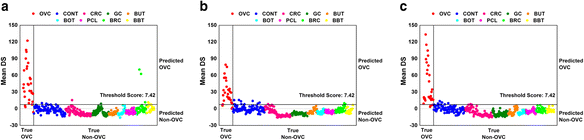
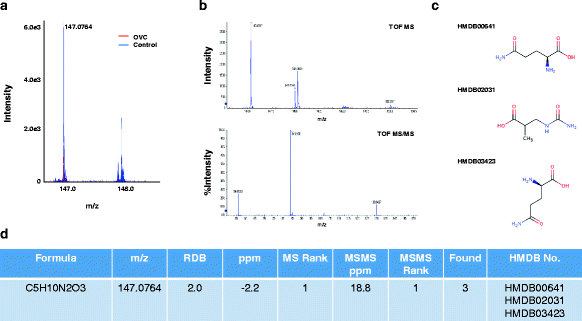
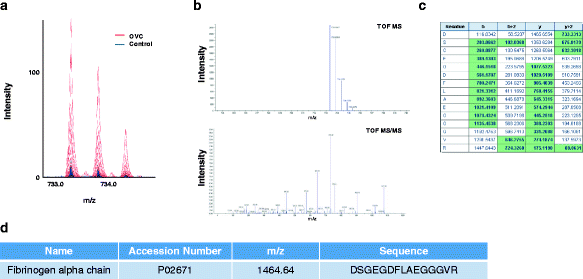
Results: Matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry was performed to obtain mass spectral data on metabolites detected as LMIs up to a mass-to-charge ratio (m/z) of 2500 for 1184 serum samples collected from healthy individuals and patients with OVC, other types of cancer, or several types of benign tumor. Principal component analysis-based discriminant analysis and two search algorithms were employed to identify discriminative low-mass ions for distinguishing OVC from non-OVC cases. OVC LOME with 13 discriminative LMIs produced excellent classification results in a validation set (sensitivity, 93.10 %; specificity, 100.0 %). Among 13 LMIs showing differential mass intensities in OVC, 3 metabolic compounds were identified and semi-quantitated. The relative amount of LPC 16:0 was somewhat decreased in OVC, but not significantly so. In contrast, D,L-glutamine and fibrinogen alpha chain fragment were significantly increased in OVC compared to the control group (p = 0.001 and 0.002, respectively).
Conclusion: The present study suggested that OVC LOME might be a useful non-invasive tool with high sensitivity and specificity for OVC screening. The LOME approach could enable screening for multiple diseases, including various types of cancer, based on a single blood sample. Furthermore, the serum levels of three metabolic compounds-D,L-glutamine, LPC 16:0 and fibrinogen alpha chain fragment-might facilitate screening for OVC.
Keywords: MALDI-TOF mass spectrometry; Ovarian cancer; Pattern recognition; Screening; Serum profiling.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources

