Diversity of plasmids and Tn1546-type transposons among VanA Enterococcus faecium in Poland
- PMID: 27752789
- PMCID: PMC5253160
- DOI: 10.1007/s10096-016-2804-8
Diversity of plasmids and Tn1546-type transposons among VanA Enterococcus faecium in Poland
Abstract
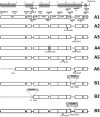
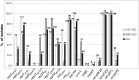
The objective of this study was to investigate the antimicrobial resistance, Tn1546 transposon variability and plasmid diversity among Polish vancomycin-resistant Enterococcus faecium (VREfm) isolates of VanA phenotype in the context of their clonal structure. Two hundred sixteen clinical VREfm isolates collected between 1997 and 2010 were studied by antimicrobial susceptibility testing, MLST, MLVA and detection of IS16, esp Efm, pilA, intA and plasmid-specific genes by PCR. Tn1546 structure was revealed by overlapping PCR and sequencing. Selected isolates were subjected to PFGE-S1 and Southern hybridization analyses. The vast majority of the isolates (95.8 %) belonged to lineages 17/18 (during the whole study period 1997-2010) and 78 (mostly in 2006-2010) of hospital-adapted meroclone of E. faecium. All isolates displayed a multi-drug resistance phenotype. Twenty-eight Tn1546 types (including 26 novel ones) were associated with eight different ISs (IS1216, IS1251, ISEfa4, ISEfa5, ISEfm2, ISEf1, IS3-like, ISEfm1-like). The vanA-determinant was typically located on plasmids, which most commonly carried rep2pRE25, rep17pRUM, rep18pEF418, rep1pIP501, ω-ε-ζ and axe-txe genes. VanA isolates from 1997-2005 to 2006-2010 differed in clonal composition, prevalence of gentamicin- and tetracycline-resistance and plasmidome. Our analysis revealed high complexity of Tn1546-type transposons and vanA-plasmids, and suggested that diverse genetic events, such as conjugation transfer, recombination, chromosomal integration and DNA mutations shaped the structure of these elements among Polish VREfm.
Conflict of interest statement
Compliance with ethical standards Funding This work was supported by the grant N N401588540 from the Narodowe Centrum Nauki (NCN), Poland, by the MIKROBANK funding from the Ministry of Science and Higher Education, Poland, and by a statutory funding from the Ministry of Science and Higher Education, Poland. Conflict of interest The authors declare that they have no conflict of interest. Ethical approval and informed consent Isolates were obtained as a part of routine activity of the NRCST and were analysed anonymously in a retrospective manner. Ethical approval and informed consent were thus not required.
Figures



Similar articles
-
VanA-Enterococcus faecalis in Poland: hospital population clonal structure and vanA mobilome.Eur J Clin Microbiol Infect Dis. 2022 Oct;41(10):1245-1261. doi: 10.1007/s10096-022-04479-4. Epub 2022 Sep 3. Eur J Clin Microbiol Infect Dis. 2022. PMID: 36057762 Free PMC article.
-
Genomic analysis of 495 vancomycin-resistant Enterococcus faecium reveals broad dissemination of a vanA plasmid in more than 19 clones from Copenhagen, Denmark.J Antimicrob Chemother. 2017 Jan;72(1):40-47. doi: 10.1093/jac/dkw360. Epub 2016 Sep 7. J Antimicrob Chemother. 2017. PMID: 27605596
-
Multilevel population genetic analysis of vanA and vanB Enterococcus faecium causing nosocomial outbreaks in 27 countries (1986-2012).J Antimicrob Chemother. 2016 Dec;71(12):3351-3366. doi: 10.1093/jac/dkw312. Epub 2016 Aug 15. J Antimicrob Chemother. 2016. PMID: 27530756
-
Strategies and approaches in plasmidome studies-uncovering plasmid diversity disregarding of linear elements?Front Microbiol. 2015 May 26;6:463. doi: 10.3389/fmicb.2015.00463. eCollection 2015. Front Microbiol. 2015. PMID: 26074886 Free PMC article. Review.
-
Antimicrobial Chemotherapy in Japan: A Historical Look.J Infect Chemother. 1996;1(3):153-165. doi: 10.1007/BF02350643. Epub 2014 Apr 5. J Infect Chemother. 1996. PMID: 29681358 Review. No abstract available.
Cited by
-
Antimicrobial Resistance Gene Detection and Plasmid Typing Among Multidrug Resistant Enterococci Isolated from Freshwater Environment.Microorganisms. 2020 Sep 2;8(9):1338. doi: 10.3390/microorganisms8091338. Microorganisms. 2020. PMID: 32887339 Free PMC article.
-
Characterization of a Tigecycline-, Linezolid- and Vancomycin-Resistant Clinical Enteroccoccus faecium Isolate, Carrying vanA and vanB Genes.Infect Dis Ther. 2023 Nov;12(11):2545-2565. doi: 10.1007/s40121-023-00881-3. Epub 2023 Oct 11. Infect Dis Ther. 2023. PMID: 37821741 Free PMC article.
-
Isolation and characterization of a multidrug-resistant Clostridioides difficile toxinotype V from municipal wastewater treatment plant.J Environ Health Sci Eng. 2020 Sep 26;18(2):1281-1288. doi: 10.1007/s40201-020-00546-0. eCollection 2020 Dec. J Environ Health Sci Eng. 2020. PMID: 33312642 Free PMC article.
-
Genetic Basis of Emerging Vancomycin, Linezolid, and Daptomycin Heteroresistance in a Case of Persistent Enterococcus faecium Bacteremia.Antimicrob Agents Chemother. 2018 Mar 27;62(4):e02007-17. doi: 10.1128/AAC.02007-17. Print 2018 Apr. Antimicrob Agents Chemother. 2018. PMID: 29339387 Free PMC article.
-
VanA-Enterococcus faecalis in Poland: hospital population clonal structure and vanA mobilome.Eur J Clin Microbiol Infect Dis. 2022 Oct;41(10):1245-1261. doi: 10.1007/s10096-022-04479-4. Epub 2022 Sep 3. Eur J Clin Microbiol Infect Dis. 2022. PMID: 36057762 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

