Using Chemical Probes to Assess the Feasibility of Targeting SecA for Developing Antimicrobial Agents against Gram-Negative Bacteria
- PMID: 27753464
- PMCID: PMC5189635
- DOI: 10.1002/cmdc.201600421
Using Chemical Probes to Assess the Feasibility of Targeting SecA for Developing Antimicrobial Agents against Gram-Negative Bacteria
Abstract
With the widespread emergence of drug resistance, there is an urgent need to search for new antimicrobials, especially those against Gram-negative bacteria. Along this line, the identification of viable targets is a critical first step. The protein translocase SecA is commonly believed to be an excellent target for the development of broad-spectrum antimicrobials. In recent years, we developed three structural classes of SecA inhibitors that have proven to be very effective against Gram-positive bacteria. However, we have not achieved the same level of success against Gram-negative bacteria, despite the potent inhibition of SecA in enzyme assays by the same inhibitors. In this study, we use representative inhibitors as chemical probes to gain an understanding as to why these inhibitors were not effective against Gram-negative bacteria. The results validate our initial postulation that the major difference in effectiveness against Gram-positive and Gram-negative bacteria is in the additional permeability barrier posed by the outer membrane of Gram-negative bacteria. We also found that the expression of efflux pumps, which are responsible for multidrug resistance (MDR), have no effect on the effectiveness of these SecA inhibitors. Identification of an inhibitor-resistant mutant and complementation tests of the plasmids containing secA in a secAts mutant showed that a single secA-azi-9 mutation increased the resistance, providing genetic evidence that SecA is indeed the target of these inhibitors in bacteria. Such results strongly suggest SecA as an excellent target for developing effective antimicrobials against Gram-negative bacteria with the intrinsic ability to overcome MDR. A key future research direction should be the optimization of membrane permeability.
Keywords: Gram-negative bacteria; SecA translocase; antibiotics; membrane permeability; multidrug resistance.
© 2016 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures

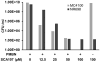
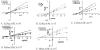


Similar articles
-
SecA inhibitors as potential antimicrobial agents: differential actions on SecA-only and SecA-SecYEG protein-conducting channels.FEMS Microbiol Lett. 2018 Aug 1;365(15):fny145. doi: 10.1093/femsle/fny145. FEMS Microbiol Lett. 2018. PMID: 30007321 Free PMC article. Review.
-
Evaluation of small molecule SecA inhibitors against methicillin-resistant Staphylococcus aureus.Bioorg Med Chem. 2015 Nov 1;23(21):7061-8. doi: 10.1016/j.bmc.2015.09.027. Epub 2015 Sep 21. Bioorg Med Chem. 2015. PMID: 26432604 Free PMC article.
-
Design, Synthesis and Evaluation of Triazole-Pyrimidine Analogues as SecA Inhibitors.ChemMedChem. 2016 Jan 5;11(1):43-56. doi: 10.1002/cmdc.201500447. Epub 2015 Nov 26. ChemMedChem. 2016. PMID: 26607404 Free PMC article.
-
SecA: a potential antimicrobial target.Future Med Chem. 2015;7(8):989-1007. doi: 10.4155/fmc.15.42. Future Med Chem. 2015. PMID: 26062397 Free PMC article. Review.
-
Fluorescein analogues inhibit SecA ATPase: the first sub-micromolar inhibitor of bacterial protein translocation.ChemMedChem. 2012 Apr;7(4):571-7. doi: 10.1002/cmdc.201100594. Epub 2012 Feb 22. ChemMedChem. 2012. PMID: 22354575 Free PMC article.
Cited by
-
The Dynamic SecYEG Translocon.Front Mol Biosci. 2021 Apr 15;8:664241. doi: 10.3389/fmolb.2021.664241. eCollection 2021. Front Mol Biosci. 2021. PMID: 33937339 Free PMC article. Review.
-
Inhibitors of protein translocation across membranes of the secretory pathway: novel antimicrobial and anticancer agents.Cell Mol Life Sci. 2018 May;75(9):1541-1558. doi: 10.1007/s00018-017-2743-2. Epub 2018 Jan 5. Cell Mol Life Sci. 2018. PMID: 29305616 Free PMC article. Review.
-
Unrealized targets in the discovery of antibiotics for Gram-negative bacterial infections.Nat Rev Drug Discov. 2023 Dec;22(12):957-975. doi: 10.1038/s41573-023-00791-6. Epub 2023 Oct 13. Nat Rev Drug Discov. 2023. PMID: 37833553 Review.
-
SecA inhibitors as potential antimicrobial agents: differential actions on SecA-only and SecA-SecYEG protein-conducting channels.FEMS Microbiol Lett. 2018 Aug 1;365(15):fny145. doi: 10.1093/femsle/fny145. FEMS Microbiol Lett. 2018. PMID: 30007321 Free PMC article. Review.
-
Permeability barriers of Gram-negative pathogens.Ann N Y Acad Sci. 2020 Jan;1459(1):5-18. doi: 10.1111/nyas.14134. Epub 2019 Jun 4. Ann N Y Acad Sci. 2020. PMID: 31165502 Free PMC article. Review.
References
-
- Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Clin Infect Dis. 2009;48:1–12. - PubMed
- Bonomo RA, Szabo D. Clin Infect Dis. 2006;43(Suppl 2):S49–56. - PubMed
- Chopra I, Schofield C, Everett M, O’Neill A, Miller K, Wilcox M, Frere JM, Dawson M, Czaplewski L, Urleb U, Courvalin P. Lancet Infect Dis. 2008;8:133–139. - PubMed
- Cornaglia G, Rossolini GM. Clin Microbiol Infect. 2010;16:99–101. - PubMed
- Livermore DM. J Antimicrob Chemother. 2009;64(Suppl 1):i29–36. - PubMed
-
- Nakane A, Takamatsu H, Oguro A, Sadaie Y, Nakamura K, Yamane K. Microbiology. 1995;141(Pt 1):113–121. - PubMed
- Nathan C. Nature. 2004;431:899–902. - PubMed
- Nikaido H. Microbiol Mol Biol Rev. 2003;67:593–656. - PMC - PubMed
- Nikaido H. Annu Rev Biochem. 2009;78:119–146. - PMC - PubMed
- Nikaido H, Pages JM. FEMS Microbiol Rev. 2012;36:340–363. - PMC - PubMed
- Nikaido H, Zgurskaya HI. Curr Opin Infect Dis. 1999;12:529–536. - PubMed
- Oliver DB, Cabelli RJ, Dolan KM, Jarosik GP. Proc Natl Acad Sci USA. 1990;87:8227–8231. - PMC - PubMed
- Osborne RS, Silhavy TJ. EMBO J. 1993;12:3391–3398. - PMC - PubMed
-
- Chaudhary AS, Chen W, Jin J, Tai PC, Wang B. Future Med Chem. 2015;7:989–1007. - PMC - PubMed
- Evans ME, Feola DJ, Rapp RP. Ann Pharmacother. 1999;33:960–967. - PubMed
- Lin BR, Hsieh YH, Jiang C, Tai PC. The Journal of membrane biology. 2012;245:747–757. - PubMed
- Rao S, De Waelheyns CVE, Economou A, Anné J. Biochimi Biophys Acta (BBA) - Mol Cell Res. 2014;1843:1762–1783. - PubMed
-
- Jang MY, De Jonghe S, Segers K, Anne J, Herdewijn P. Bioorg Med Chem. 2011;19:702–714. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

