Golgi proteins in circulating human platelets are distributed across non-stacked, scattered structures
- PMID: 27753523
- PMCID: PMC5723161
- DOI: 10.1080/09537104.2016.1235685
Golgi proteins in circulating human platelets are distributed across non-stacked, scattered structures
Abstract
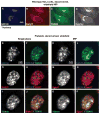
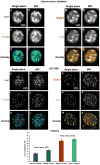
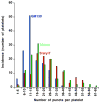
Platelets are small, anucleate cell fragments that are central to hemostasis, thrombosis, and inflammation. They are derived from megakaryocytes from which they inherit their organelles. As platelets can synthesize proteins and contain many of the enzymes of the secretory pathway, one might expect all mature human platelets to contain a stacked Golgi apparatus, the central organelle of the secretory pathway. By thin section electron microscopy, stacked membranes resembling the stacked Golgi compartment in megakaryocytes and other nucleated cells can be detected in both proplatelets and platelets. However, the incidence of such structures is low and whether each and every platelet contains such a structure remains an open question. By single-label, immunofluorescence staining, Golgi glycosyltransferases are found within each platelet and map to scattered structures. Whether these structures are positive for marker proteins from multiple Golgi subcompartments remains unknown. Here, we have applied state-of-the-art techniques to probe the organization state of the Golgi apparatus in resting human platelets. By the whole cell volume technique of serial-block-face scanning electron microscopy (SBF-SEM), we failed to observe stacked, Golgi-like structures in any of the 65 platelets scored. When antibodies directed against Golgi proteins were tested against HeLa cells, labeling was restricted to an elongated juxtanuclear ribbon characteristic of a stacked Golgi apparatus. By multi-label immunofluorescence microscopy, we found that each and every resting human platelet was positive for cis, trans, and trans Golgi network (TGN) proteins. However, in each case, the proteins were found in small puncta scattered about the platelet. At the resolution of deconvolved, widefield fluorescence microscopy, these proteins had limited tendency to map adjacent to one another. When the results of 3D structured illumination microscopy (3D SIM), a super resolution technique, were scored quantitatively, the Golgi marker proteins failed to map together indicating at the protein level considerable degeneration of the platelet Golgi apparatus relative to the layered stack as seen in the megakaryocyte. In conclusion, we suggest that these results have important implications for organelle structure/function relationships in the mature platelet and the extent to which Golgi apparatus organization is maintained in platelets. Our results suggest that Golgi proteins in circulating platelets are present within a series of scattered, separated structures. As separate elements, selective sets of Golgi enzymes or sugar nucleotides could be secreted during platelet activation. The establishment of the functional importance, if any, of these scattered structures in sequential protein modification in circulating platelets will require further research.
Keywords: 3D SIM; Golgi apparatus; fluorescence microscopy; platelets; serial-block-face scanning electron microscopy.
Conflict of interest statement
This work was supported in part by NIH grants S10 OD018065, R01 HL119393, and R01 GM092960. Work in the Leapman laboratory was supported by the NIBIB, NIH intramural program. The authors report no other declarations of interest.
Figures


Similar articles
-
Scattered Golgi elements during microtubule disruption are initially enriched in trans-Golgi proteins.Mol Biol Cell. 1998 Jan;9(1):191-207. doi: 10.1091/mbc.9.1.191. Mol Biol Cell. 1998. PMID: 9437000 Free PMC article.
-
Three-dimensional shape of the Golgi apparatus in different cell types: serial section scanning electron microscopy of the osmium-impregnated Golgi apparatus.Microscopy (Oxf). 2016 Apr;65(2):145-57. doi: 10.1093/jmicro/dfv360. Epub 2015 Nov 24. Microscopy (Oxf). 2016. PMID: 26609075
-
Golgi complexes in hypogranular platelet syndromes.Platelets. 2005 Feb;16(1):51-60. doi: 10.1080/0953710042000260173. Platelets. 2005. PMID: 15763897
-
The study of the Golgi apparatus in blood--basic science and clinical applications.Clin Lab. 2010;56(5-6):231-43. Clin Lab. 2010. PMID: 20575472 Review.
-
Novel scanning electron microscopy methods for analyzing the 3D structure of the Golgi apparatus.Anat Sci Int. 2017 Jan;92(1):37-49. doi: 10.1007/s12565-016-0380-8. Epub 2016 Oct 26. Anat Sci Int. 2017. PMID: 27785745 Review.
Cited by
-
The ins and outs of endocytic trafficking in platelet functions.Curr Opin Hematol. 2017 Sep;24(5):467-474. doi: 10.1097/MOH.0000000000000366. Curr Opin Hematol. 2017. PMID: 28650849 Free PMC article. Review.
-
A Divergent Platelet Transcriptome in Patients with Lipedema and Lymphedema.Genes (Basel). 2024 Jun 4;15(6):737. doi: 10.3390/genes15060737. Genes (Basel). 2024. PMID: 38927673 Free PMC article.
-
Canalicular system reorganization during mouse platelet activation as revealed by 3D ultrastructural analysis.Platelets. 2021 Jan 2;32(1):97-104. doi: 10.1080/09537104.2020.1719993. Epub 2020 Jan 31. Platelets. 2021. PMID: 32000578 Free PMC article.
-
Impact of Escherichia coli K12 and O18:K1 on human platelets: Differential effects on platelet activation, RNAs and proteins.Sci Rep. 2018 Nov 1;8(1):16145. doi: 10.1038/s41598-018-34473-w. Sci Rep. 2018. PMID: 30385858 Free PMC article.
-
3D ultrastructural analysis of α-granule, dense granule, mitochondria, and canalicular system arrangement in resting human platelets.Res Pract Thromb Haemost. 2019 Oct 25;4(1):72-85. doi: 10.1002/rth2.12260. eCollection 2020 Jan. Res Pract Thromb Haemost. 2019. PMID: 31989087 Free PMC article.
References
-
- King S, Reed GL. Development of platelet secretory granules. Semin Cell Dev Biol. 2002;13:203–302. - PubMed
-
- Warshaw AL, Laster L, Shulman NR. Protein Synthesis by Human Platelets. J Biol Chem. 1967;242(9):2094–7. - PubMed
-
- Schubert P, Devine DV. De novo protein synthesis in mature platelets: a consideration for transfusion medicine. Vox Sang. 2010;99(2):112–22. - PubMed
-
- Schwertz H, Rowley JW, Tolley ND, Campbell RA, Weyrich AS. Assessing protein synthesis by platelets. Methods Mol Biol. 2012;788:141–53. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
