Dysfunctional mitochondrial fission impairs cell reprogramming
- PMID: 27753531
- PMCID: PMC5176137
- DOI: 10.1080/15384101.2016.1241930
Dysfunctional mitochondrial fission impairs cell reprogramming
Abstract
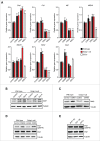
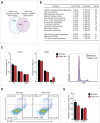
We have recently shown that mitochondrial fission is induced early in reprogramming in a Drp1-dependent manner; however, the identity of the factors controlling Drp1 recruitment to mitochondria was unexplored. To investigate this, we used a panel of RNAi targeting factors involved in the regulation of mitochondrial dynamics and we observed that MiD51, Gdap1 and, to a lesser extent, Mff were found to play key roles in this process. Cells derived from Gdap1-null mice were used to further explore the role of this factor in cell reprogramming. Microarray data revealed a prominent down-regulation of cell cycle pathways in Gdap1-null cells early in reprogramming and cell cycle profiling uncovered a G2/M growth arrest in Gdap1-null cells undergoing reprogramming. High-Content analysis showed that this growth arrest was DNA damage-independent. We propose that lack of efficient mitochondrial fission impairs cell reprogramming by interfering with cell cycle progression in a DNA damage-independent manner.
Keywords: Gdap1; cell reprogramming; iPS cells; mitochondrial fission; pluripotency.
Figures


Comment in
-
Fission for reprogramming.Cell Cycle. 2017 Jan 17;16(2):159-160. doi: 10.1080/15384101.2016.1259898. Epub 2016 Dec 8. Cell Cycle. 2017. PMID: 27929736 Free PMC article. No abstract available.
References
-
- Westermann B. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol 2010; 11:872-884; PMID:21102612; http://dx.doi.org/10.1038/nrm3013 - DOI - PubMed
-
- Chan DC. Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev Genet 2012; 46:265-287; PMID:22934639; http://dx.doi.org/10.1146/annurev-genet-110410-132529 - DOI - PubMed
-
- Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell 2012; 148:1145-1159; PMID:22424226; http://dx.doi.org/10.1016/j.cell.2012.02.035 - DOI - PMC - PubMed
-
- Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science 2012; 337:1062-1065; PMID:22936770; http://dx.doi.org/10.1126/science.1219855. - DOI - PMC - PubMed
-
- Scorrano L. Keeping mitochondria in shape: a matter of life and death. Eur J Clin Invest 2013; 43:886-893 PMID:23869410; http://dx.doi.org/10.1111/eci.12135. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
