The Role of the Equine Herpesvirus Type 1 (EHV-1) US3-Encoded Protein Kinase in Actin Reorganization and Nuclear Egress
- PMID: 27754319
- PMCID: PMC5086611
- DOI: 10.3390/v8100275
The Role of the Equine Herpesvirus Type 1 (EHV-1) US3-Encoded Protein Kinase in Actin Reorganization and Nuclear Egress
Abstract
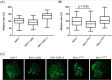
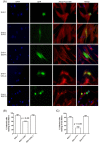
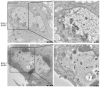
The serine-threonine protein kinase encoded by US3 gene (pUS3) of alphaherpesviruses was shown to modulate actin reorganization, cell-to-cell spread, and virus egress in a number of virus species. However, the role of the US3 orthologues of equine herpesvirus type 1 and 4 (EHV-1 and EHV-4) has not yet been studied. Here, we show that US3 is not essential for virus replication in vitro. However, growth rates and plaque diameters of a US3-deleted EHV-1 and a mutant in which the catalytic active site was destroyed were significantly reduced when compared with parental and revertant viruses or a virus in which EHV-1 US3 was replaced with the corresponding EHV-4 gene. The reduced plaque sizes were consistent with accumulation of primarily enveloped virions in the perinuclear space of the US3-negative EHV-1, a phenotype that was also rescued by the EHV-4 orthologue. Furthermore, actin stress fiber disassembly was significantly more pronounced in cells infected with parental EHV-1, revertant, or the recombinant EHV-1 expressing EHV-4 US3. Finally, we observed that deletion of US3 in EHV-1 did not affect the expression of adhesion molecules on the surface of infected cells.
Keywords: EHV-1; EHV-4; EM; US3; actin cytoskeleton; adhesion molecules.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
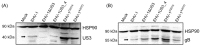



Similar articles
-
The protein encoded by the US3 orthologue of Marek's disease virus is required for efficient de-envelopment of perinuclear virions and involved in actin stress fiber breakdown.J Virol. 2005 Apr;79(7):3987-97. doi: 10.1128/JVI.79.7.3987-3997.2005. J Virol. 2005. PMID: 15767401 Free PMC article.
-
Characterization of a Herpes Simplex Virus 1 (HSV-1) Chimera in Which the Us3 Protein Kinase Gene Is Replaced with the HSV-2 Us3 Gene.J Virol. 2015 Oct 21;90(1):457-73. doi: 10.1128/JVI.02376-15. Print 2016 Jan 1. J Virol. 2015. PMID: 26491159 Free PMC article.
-
Role of gB and pUS3 in Equine Herpesvirus 1 Transfer between Peripheral Blood Mononuclear Cells and Endothelial Cells: a Dynamic In Vitro Model.J Virol. 2015 Dec;89(23):11899-908. doi: 10.1128/JVI.01809-15. Epub 2015 Sep 16. J Virol. 2015. PMID: 26378176 Free PMC article.
-
Us3 Protein Kinase Encoded by HSV: The Precise Function and Mechanism on Viral Life Cycle.Adv Exp Med Biol. 2018;1045:45-62. doi: 10.1007/978-981-10-7230-7_3. Adv Exp Med Biol. 2018. PMID: 29896662 Review.
-
Keep it in the subfamily: the conserved alphaherpesvirus US3 protein kinase.J Gen Virol. 2011 Jan;92(Pt 1):18-30. doi: 10.1099/vir.0.025593-0. Epub 2010 Oct 13. J Gen Virol. 2011. PMID: 20943887 Review.
Cited by
-
Equid Herpesvirus-1 Exploits the Extracellular Matrix of Mononuclear Cells to Ensure Transport to Target Cells.iScience. 2020 Oct 23;23(10):101615. doi: 10.1016/j.isci.2020.101615. Epub 2020 Sep 28. iScience. 2020. PMID: 33015592 Free PMC article.
-
Equine Herpesvirus Type 1 Modulates Cytokine and Chemokine Profiles of Mononuclear Cells for Efficient Dissemination to Target Organs.Viruses. 2020 Sep 8;12(9):999. doi: 10.3390/v12090999. Viruses. 2020. PMID: 32911663 Free PMC article.
-
Equine Herpesvirus 1 Bridles T Lymphocytes To Reach Its Target Organs.J Virol. 2019 Mar 21;93(7):e02098-18. doi: 10.1128/JVI.02098-18. Print 2019 Apr 1. J Virol. 2019. PMID: 30651370 Free PMC article.
-
Cytoskeletons in the Closet-Subversion in Alphaherpesvirus Infections.Viruses. 2018 Feb 13;10(2):79. doi: 10.3390/v10020079. Viruses. 2018. PMID: 29438303 Free PMC article. Review.
-
Differentiating the Roles of UL16, UL21, and Us3 in the Nuclear Egress of Herpes Simplex Virus Capsids.J Virol. 2020 Jun 16;94(13):e00738-20. doi: 10.1128/JVI.00738-20. Print 2020 Jun 16. J Virol. 2020. PMID: 32321804 Free PMC article.
References
-
- Colle C.F., 3rd, Flowers C.C., O’Callaghan D.J. Open reading frames encoding a protein kinase, homolog of glycoprotein gX of pseudorabies virus, and a novel glycoprotein map within the unique short segment of equine herpesvirus type 1. Virology. 1992;188:545–557. doi: 10.1016/0042-6822(92)90509-N. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

