Single Amino Acid Substitution N659D in HIV-2 Envelope Glycoprotein (Env) Impairs Viral Release and Hampers BST-2 Antagonism
- PMID: 27754450
- PMCID: PMC5086617
- DOI: 10.3390/v8100285
Single Amino Acid Substitution N659D in HIV-2 Envelope Glycoprotein (Env) Impairs Viral Release and Hampers BST-2 Antagonism
Abstract
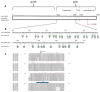
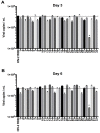
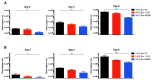
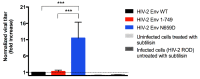
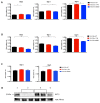
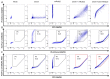
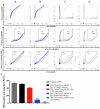
BST-2 or tetherin is a host cell restriction factor that prevents the budding of enveloped viruses at the cell surface, thus impairing the viral spread. Several countermeasures to evade this antiviral factor have been positively selected in retroviruses: the human immunodeficiency virus type 2 (HIV-2) relies on the envelope glycoprotein (Env) to overcome BST-2 restriction. The Env gp36 ectodomain seems involved in this anti-tetherin activity, however residues and regions interacting with BST-2 are not clearly defined. Among 32 HIV-2 ROD Env mutants tested, we demonstrated that the asparagine residue at position 659 located in the gp36 ectodomain is mandatory to exert the anti-tetherin function. Viral release assays in cell lines expressing BST-2 showed a loss of viral release ability for the HIV-2 N659D mutant virus compared to the HIV-2 wild type virus. In bst-2 inactivated H9 cells, those differences were lost. Subtilisin treatment of infected cells demonstrated that the N659D mutant was more tethered at the cell surface. Förster resonance energy transfer (FRET) experiments confirmed a direct molecular link between Env and BST-2 and highlighted an inability of the mutant to bind BST-2. We also tested a virus presenting a truncation of 109 amino acids at the C-terminal part of Env, a cytoplasmic tail partial deletion that is spontaneously selected in vitro. Interestingly, viral release assays and FRET experiments indicated that a full Env cytoplasmic tail was essential in BST-2 antagonism. In HIV-2 infected cells, an efficient Env-mediated antagonism of BST-2 is operated through an intermolecular link involving the asparagine 659 residue as well as the C-terminal part of the cytoplasmic tail.
Keywords: BST-2; CRISPR/Cas9 knockout; Env; FRET; HIV-2; envelope glycoprotein; gp36; restriction factors; tetherin; viral antagonist.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Preadaptation of Simian Immunodeficiency Virus SIVsmm Facilitated Env-Mediated Counteraction of Human Tetherin by Human Immunodeficiency Virus Type 2.J Virol. 2018 Aug 29;92(18):e00276-18. doi: 10.1128/JVI.00276-18. Print 2018 Sep 15. J Virol. 2018. PMID: 29976668 Free PMC article.
-
Antagonism of BST-2/Tetherin Is a Conserved Function of the Env Glycoprotein of Primary HIV-2 Isolates.J Virol. 2016 Nov 28;90(24):11062-11074. doi: 10.1128/JVI.01451-16. Print 2016 Dec 15. J Virol. 2016. PMID: 27681141 Free PMC article.
-
Determinants in HIV-2 Env and tetherin required for functional interaction.Retrovirology. 2015 Aug 7;12:67. doi: 10.1186/s12977-015-0194-0. Retrovirology. 2015. PMID: 26248668 Free PMC article.
-
[Inhibition of HBV Release by BST-2].Bing Du Xue Bao. 2016 Mar;32(2):215-21. Bing Du Xue Bao. 2016. PMID: 27396167 Review. Chinese.
-
Limiting Respiratory Viral Infection by Targeting Antiviral and Immunological Functions of BST-2/Tetherin: Knowledge and Gaps.Bioessays. 2018 Oct;40(10):e1800086. doi: 10.1002/bies.201800086. Epub 2018 Aug 16. Bioessays. 2018. PMID: 30113067 Free PMC article. Review.
Cited by
-
CRISPR/Cas9-Advancing Orthopoxvirus Genome Editing for Vaccine and Vector Development.Viruses. 2018 Jan 22;10(1):50. doi: 10.3390/v10010050. Viruses. 2018. PMID: 29361752 Free PMC article. Review.
-
Immunity and Viral Infections: Modulating Antiviral Response via CRISPR-Cas Systems.Viruses. 2021 Jul 15;13(7):1373. doi: 10.3390/v13071373. Viruses. 2021. PMID: 34372578 Free PMC article. Review.
-
Preadaptation of Simian Immunodeficiency Virus SIVsmm Facilitated Env-Mediated Counteraction of Human Tetherin by Human Immunodeficiency Virus Type 2.J Virol. 2018 Aug 29;92(18):e00276-18. doi: 10.1128/JVI.00276-18. Print 2018 Sep 15. J Virol. 2018. PMID: 29976668 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

