Glycosphingolipid-Protein Interaction in Signal Transduction
- PMID: 27754465
- PMCID: PMC5085762
- DOI: 10.3390/ijms17101732
Glycosphingolipid-Protein Interaction in Signal Transduction
Abstract
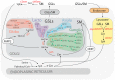
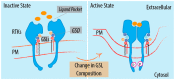
Glycosphingolipids (GSLs) are a class of ceramide-based glycolipids essential for embryo development in mammals. The synthesis of specific GSLs depends on the expression of distinctive sets of GSL synthesizing enzymes that is tightly regulated during development. Several reports have described how cell surface receptors can be kept in a resting state or activate alternative signalling events as a consequence of their interaction with GSLs. Specific GSLs, indeed, interface with specific protein domains that are found in signalling molecules and which act as GSL sensors to modify signalling responses. The regulation exerted by GSLs on signal transduction is orthogonal to the ligand-receptor axis, as it usually does not directly interfere with the ligand binding to receptors. Due to their properties of adjustable production and orthogonal action on receptors, GSLs add a new dimension to the control of the signalling in development. GSLs can, indeed, dynamically influence progenitor cell response to morphogenetic stimuli, resulting in alternative differentiation fates. Here, we review the available literature on GSL-protein interactions and their effects on cell signalling and development.
Keywords: glycan–protein interaction; glycosphingolipid; signalling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Lindquist S., Karitkina D., Langnaese K., Posevitz-Fejfar A., Schraven B., Xavier R., Seed B., Lindquist J.A. Phosphoprotein associated with glycosphingolipid-enriched microdomains differentially modulates SRC kinase activity in brain maturation. PLoS ONE. 2011;6:1732. doi: 10.1371/journal.pone.0023978. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

