Fbxo28 promotes mitotic progression and regulates topoisomerase IIα-dependent DNA decatenation
- PMID: 27754753
- PMCID: PMC5224455
- DOI: 10.1080/15384101.2016.1246093
Fbxo28 promotes mitotic progression and regulates topoisomerase IIα-dependent DNA decatenation
Abstract
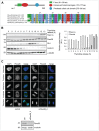
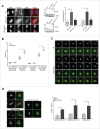
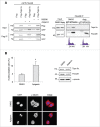
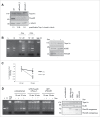
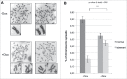
Topoisomerase IIα is an essential enzyme that resolves topological constraints in genomic DNA. It functions in disentangling intertwined chromosomes during anaphase leading to chromosome segregation thus preserving genomic stability. Here we describe a previously unrecognized mechanism regulating topoisomerase IIα activity that is dependent on the F-box protein Fbxo28. We find that Fbxo28, an evolutionarily conserved protein, is required for proper mitotic progression. Interfering with Fbxo28 function leads to a delay in metaphase-to-anaphase progression resulting in mitotic defects as lagging chromosomes, multipolar spindles and multinucleation. Furthermore, we find that Fbxo28 interacts and colocalizes with topoisomerase IIα throughout the cell cycle. Depletion of Fbxo28 results in an increase in topoisomerase IIα-dependent DNA decatenation activity. Interestingly, blocking the interaction between Fbxo28 and topoisomerase IIα also results in multinucleated cells. Our findings suggest that Fbxo28 regulates topoisomerase IIα decatenation activity and plays an important role in maintaining genomic stability.
Keywords: Cell cycle; F-box protein; Fbxo28; SCF; Topoisomerase IIα; decatenation; mitosis.
Figures
References
-
- Chen SH, Chan NL, Hsieh TS. New mechanistic and functional insights into DNA topoisomerases. Annu Rev Biochem 2013; 82:139-70; PMID:23495937; http://dx.doi.org/10.1146/annurev-biochem-061809-100002 - DOI - PubMed
-
- Heck MM, Hittelman WN, Earnshaw WC. Differential expression of DNA topoisomerases I and II during the eukaryotic cell cycle. Proc Natl Acad Sci U S A 1988; 85:1086-90; PMID:2829215; http://dx.doi.org/10.1073/pnas.85.4.1086 - DOI - PMC - PubMed
-
- Ishida R, Sato M, Narita T, Utsumi KR, Nishimoto T, Morita T, Nagata H, Andoh T. Inhibition of DNA topoisomerase II by ICRF-193 induces polyploidization by uncoupling chromosome dynamics from other cell cycle events. J Cell Biol 1994; 126:1341-51; PMID:8089169; http://dx.doi.org/10.1083/jcb.126.6.1341 - DOI - PMC - PubMed
-
- Oliveira RA, Hamilton RS, Pauli A, Davis I, Nasmyth K. Cohesin cleavage and Cdk inhibition trigger formation of daughter nuclei. Nat Cell Biol 2010; 12:185-92; PMID:20081838; http://dx.doi.org/10.1038/ncb2018 - DOI - PMC - PubMed
-
- Holm C, Goto T, Wang JC, Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell 1985; 41:553-63; PMID:2985283; http://dx.doi.org/10.1016/S0092-8674(85)80028-3 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
