Muscadine Grape Skin Extract Induces an Unfolded Protein Response-Mediated Autophagy in Prostate Cancer Cells: A TMT-Based Quantitative Proteomic Analysis
- PMID: 27755556
- PMCID: PMC5068743
- DOI: 10.1371/journal.pone.0164115
Muscadine Grape Skin Extract Induces an Unfolded Protein Response-Mediated Autophagy in Prostate Cancer Cells: A TMT-Based Quantitative Proteomic Analysis
Abstract
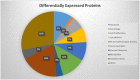
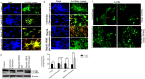
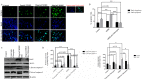
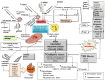
Muscadine grape skin extract (MSKE) is derived from muscadine grape (Vitis rotundifolia), a common red grape used to produce red wine. Endoplasmic reticulum (ER) stress activates the unfolded protein response (UPR) that serves as a survival mechanism to relieve ER stress and restore ER homeostasis. However, when persistent, ER stress can alter the cytoprotective functions of the UPR to promote autophagy and cell death. Although MSKE has been documented to induce apoptosis, it has not been linked to ER stress/UPR/autophagy. We hypothesized that MSKE may induce a severe ER stress response-mediated autophagy leading to apoptosis. As a model, we treated C4-2 prostate cancer cells with MSKE and performed a quantitative Tandem Mass Tag Isobaric Labeling proteomic analysis. ER stress response, autophagy and apoptosis were analyzed by western blot, acridine orange and TUNEL/Annexin V staining, respectively. Quantitative proteomics analysis indicated that ER stress response proteins, such as GRP78 were greatly elevated following treatment with MSKE. The up-regulation of pro-apoptotic markers PARP, caspase-12, cleaved caspase-3, -7, BAX and down-regulation of anti-apoptotic marker BCL2 was confirmed by Western blot analysis and apoptosis was visualized by increased TUNEL/Annexin V staining upon MSKE treatment. Moreover, increased acridine orange, and LC3B staining was detected in MSKE-treated cells, suggesting an ER stress/autophagy response. Finally, MSKE-mediated autophagy and apoptosis was antagonized by co-treatment with chloroquine, an autophagy inhibitor. Our results indicate that MSKE can elicit an UPR that can eventually lead to apoptosis in prostate cancer cells.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Targeting multiple pro-apoptotic signaling pathways with curcumin in prostate cancer cells.PLoS One. 2017 Jun 19;12(6):e0179587. doi: 10.1371/journal.pone.0179587. eCollection 2017. PLoS One. 2017. PMID: 28628644 Free PMC article.
-
Muscadine grape skin extract reverts snail-mediated epithelial mesenchymal transition via superoxide species in human prostate cancer cells.BMC Complement Altern Med. 2014 Mar 12;14:97. doi: 10.1186/1472-6882-14-97. BMC Complement Altern Med. 2014. PMID: 24617993 Free PMC article.
-
CCAAT-displacement protein/cut homeobox transcription factor (CUX1) represses estrogen receptor-alpha (ER-α) in triple-negative breast cancer cells and can be antagonized by muscadine grape skin extract (MSKE).PLoS One. 2019 Apr 9;14(4):e0214844. doi: 10.1371/journal.pone.0214844. eCollection 2019. PLoS One. 2019. PMID: 30964885 Free PMC article.
-
Phytochemical-mediated modulation of autophagy and endoplasmic reticulum stress as a cancer therapeutic approach.Phytother Res. 2024 Sep;38(9):4353-4385. doi: 10.1002/ptr.8283. Epub 2024 Jul 3. Phytother Res. 2024. PMID: 38961675 Review.
-
Hyperthermia and protein homeostasis: Cytoprotection and cell death.J Therm Biol. 2020 Jul;91:102615. doi: 10.1016/j.jtherbio.2020.102615. Epub 2020 May 8. J Therm Biol. 2020. PMID: 32716865 Review.
Cited by
-
Oncogenic Proteomics Approaches for Translational Research and HIV-Associated Malignancy Mechanisms.Proteomes. 2023 Jul 4;11(3):22. doi: 10.3390/proteomes11030022. Proteomes. 2023. PMID: 37489388 Free PMC article. Review.
-
Proteomics-Metabolomics Combined Approach Identifies Peroxidasin as a Protector against Metabolic and Oxidative Stress in Prostate Cancer.Int J Mol Sci. 2019 Jun 21;20(12):3046. doi: 10.3390/ijms20123046. Int J Mol Sci. 2019. PMID: 31234468 Free PMC article.
-
Effect of Ball Mill Treatment on the Physicochemical Properties and Digestibility of Protein Extracts Generated from Scallops (Chlamys farreri).Int J Mol Sci. 2018 Feb 9;19(2):531. doi: 10.3390/ijms19020531. Int J Mol Sci. 2018. PMID: 29425186 Free PMC article.
-
Essential role of JunD in cell proliferation is mediated via MYC signaling in prostate cancer cells.Cancer Lett. 2019 Apr 28;448:155-167. doi: 10.1016/j.canlet.2019.02.005. Epub 2019 Feb 11. Cancer Lett. 2019. PMID: 30763715 Free PMC article.
-
Novel roles for HMGA2 isoforms in regulating oxidative stress and sensitizing to RSL3-Induced ferroptosis in prostate cancer cells.Heliyon. 2023 Apr 7;9(4):e14810. doi: 10.1016/j.heliyon.2023.e14810. eCollection 2023 Apr. Heliyon. 2023. PMID: 37113783 Free PMC article.
References
-
- Meiyanto E, Hermawan A, Anindyajati (2012) Natural products for cancer-targeted therapy: citrus flavonoids as potent chemopreventive agents. Asian Pac J Cancer Prev 13: 427–436. - PubMed
-
- Divisi D. DT S, Salvemini S., Garramone M., and Crisci R.. (2006) Diet and cancer. Acta Biomed 77: 118–123. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

