Characterization of Triticum aestivum Abscisic Acid Receptors and a Possible Role for These in Mediating Fusairum Head Blight Susceptibility in Wheat
- PMID: 27755583
- PMCID: PMC5068739
- DOI: 10.1371/journal.pone.0164996
Characterization of Triticum aestivum Abscisic Acid Receptors and a Possible Role for These in Mediating Fusairum Head Blight Susceptibility in Wheat
Abstract
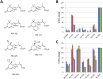
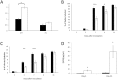
Abscisic acid (ABA) is a well-characterized plant hormone, known to mediate developmental aspects as well as both abiotic and biotic stress responses. Notably, the exogenous application of ABA has recently been shown to increase susceptibility to the fungal pathogen Fusarium graminearum, the causative agent of Fusarium head blight (FHB) in wheat and other cereals. However roles and mechanisms associated with ABA's modulation of pathogen responses remain enigmatic. Here the identification of putative ABA receptors from available genomic databases for Triticum aestivum (bread wheat) and Brachypodium distachyon (a model cereal) are reported. A number of these were cloned for recombinant expression and their functionality as ABA receptors confirmed by in vitro assays against protein phosphatases Type 2Cs. Ligand selectivity profiling of one of the wheat receptors (Ta_PYL2DS_FL) highlighted unique activities compared to Arabidopsis AtPYL5. Mutagenic analysis showed Ta_PYL2DS_FL amino acid D180 as being a critical contributor to this selectivity. Subsequently, a virus induced gene silencing (VIGS) approach was used to knockdown wheat Ta_PYL4AS_A (and similar) in planta, yielding plants with increased early stage resistance to FHB progression and decreased mycotoxin accumulation. Together these results confirm the existence of a family of ABA receptors in wheat and Brachypodium and present insight into factors modulating receptor function at the molecular level. That knockdown of Ta_PYL4AS_A (and similar) leads to early stage FHB resistance highlights novel targets for investigation in the future development of disease resistant crops.
Conflict of interest statement
Marci Surpin is employed by Valent Biosciences Corporation. The other authors confirm that no competing interests exist. There are no patents, products in development or marketed products to declare. This does not alter our adherence to all the PLOS ONE policies on sharing data and materials, as detailed online in the guide for authors.
Figures


References
-
- Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, et al. The abscisic acid receptor PYR1 in complexwith abscisic acid. Nature. 2009a;462: 665–668. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

