Mitochondrial Genomes of Kinorhyncha: trnM Duplication and New Gene Orders within Animals
- PMID: 27755612
- PMCID: PMC5068742
- DOI: 10.1371/journal.pone.0165072
Mitochondrial Genomes of Kinorhyncha: trnM Duplication and New Gene Orders within Animals
Abstract
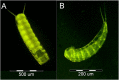
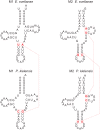
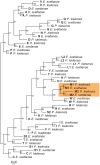
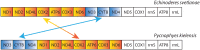
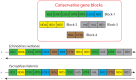
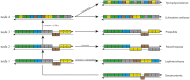
Many features of mitochondrial genomes of animals, such as patterns of gene arrangement, nucleotide content and substitution rate variation are extensively used in evolutionary and phylogenetic studies. Nearly 6,000 mitochondrial genomes of animals have already been sequenced, covering the majority of animal phyla. One of the groups that escaped mitogenome sequencing is phylum Kinorhyncha-an isolated taxon of microscopic worm-like ecdysozoans. The kinorhynchs are thought to be one of the early-branching lineages of Ecdysozoa, and their mitochondrial genomes may be important for resolving evolutionary relations between major animal taxa. Here we present the results of sequencing and analysis of mitochondrial genomes from two members of Kinorhyncha, Echinoderes svetlanae (Cyclorhagida) and Pycnophyes kielensis (Allomalorhagida). Their mitochondrial genomes are circular molecules approximately 15 Kbp in size. The kinorhynch mitochondrial gene sequences are highly divergent, which precludes accurate phylogenetic inference. The mitogenomes of both species encode a typical metazoan complement of 37 genes, which are all positioned on the major strand, but the gene order is distinct and unique among Ecdysozoa or animals as a whole. We predict four types of start codons for protein-coding genes in E. svetlanae and five in P. kielensis with a consensus DTD in single letter code. The mitochondrial genomes of E. svetlanae and P. kielensis encode duplicated methionine tRNA genes that display compensatory nucleotide substitutions. Two distant species of Kinorhyncha demonstrate similar patterns of gene arrangements in their mitogenomes. Both genomes have duplicated methionine tRNA genes; the duplication predates the divergence of two species. The kinorhynchs share a few features pertaining to gene order that align them with Priapulida. Gene order analysis reveals that gene arrangement specific of Priapulida may be ancestral for Scalidophora, Ecdysozoa, and even Protostomia.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

