Pitfalls of improperly procured adjacent non-neoplastic tissue for somatic mutation analysis using next-generation sequencing
- PMID: 27756300
- PMCID: PMC5070097
- DOI: 10.1186/s12920-016-0226-1
Pitfalls of improperly procured adjacent non-neoplastic tissue for somatic mutation analysis using next-generation sequencing
Abstract
Background: The rapid adoption of next-generation sequencing provides an efficient system for detecting somatic alterations in neoplasms. The detection of such alterations requires a matched non-neoplastic sample for adequate filtering of non-somatic events such as germline polymorphisms. Non-neoplastic tissue adjacent to the excised neoplasm is often used for this purpose as it is simultaneously collected and generally contains the same tissue type as the neoplasm. Following NGS analysis, we and others have frequently observed low-level somatic mutations in these non-neoplastic tissues, which may impose additional challenges to somatic mutation detection as it complicates germline variant filtering.
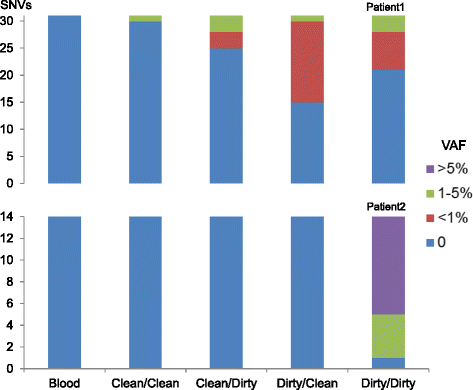
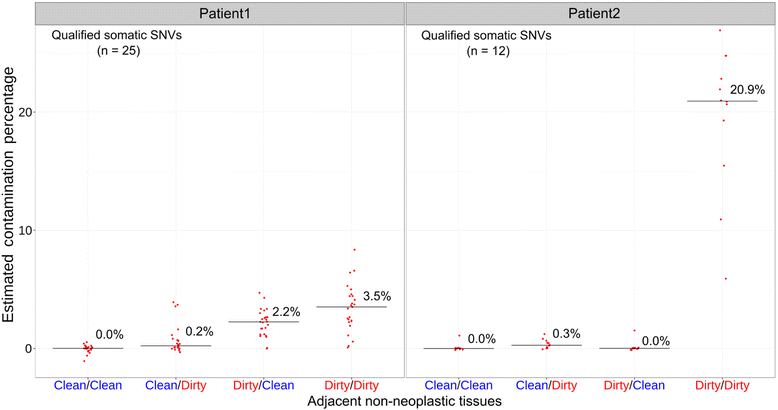
Methods: We hypothesized that the low-level somatic mutation observed in non-neoplastic tissues may be entirely or partially caused by inadvertent contamination by neoplastic cells during the surgical pathology gross assessment or tissue procurement process. To test this hypothesis, we applied a systematic protocol designed to collect multiple grossly non-neoplastic tissues using different methods surrounding each single neoplasm. The procedure was applied in two breast cancer lumpectomy specimens. In each case, all samples were first sequenced by whole-exome sequencing to identify somatic mutations in the neoplasm and determine their presence in the adjacent non-neoplastic tissues. We then generated ultra-deep coverage using targeted sequencing to assess the levels of contamination in non-neoplastic tissue samples collected under different conditions.
Results: Contamination levels in non-neoplastic tissues ranged up to 3.5 and 20.9 % respectively in the two cases tested, with consistent pattern correlated with the manner of grossing and procurement. By carefully controlling the conditions of various steps during this process, we were able to eliminate any detectable contamination in both patients.
Conclusion: The results demonstrated that the process of tissue procurement contributes to the level of contamination in non-neoplastic tissue, and contamination can be reduced to below detectable levels by using a carefully designed collection method. A standard protocol dedicated for acquiring adjacent non-neoplastic tissue that minimizes neoplasm contamination should be implemented for all somatic mutation detection studies.
Keywords: Adjacent normal tissues; Somatic mutations; Tumor contamination.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

