BcsZ inhibits biofilm phenotypes and promotes virulence by blocking cellulose production in Salmonella enterica serovar Typhimurium
- PMID: 27756305
- PMCID: PMC5070118
- DOI: 10.1186/s12934-016-0576-6
BcsZ inhibits biofilm phenotypes and promotes virulence by blocking cellulose production in Salmonella enterica serovar Typhimurium
Abstract
Background: Cellulose, a 1,4 beta-glucan polysaccharide, is produced by a variety of organisms including bacteria. Although the production of cellulose has a high biological, ecological and economical impact, regulatory mechanisms of cellulose biosynthesis are mostly unknown. Family eight cellulases are regularly associated with cellulose biosynthesis operons in bacteria; however, their function is poorly characterized. In this study, we analysed the role of the cellulase BcsZ encoded by the bcsABZC cellulose biosynthesis operon of Salmonella enterica serovar Typhimurium (S. Typhimurium) in biofilm related behavior. We also investigated the involvement of BcsZ in pathogenesis of S. Typhimurium including a murine typhoid fever infection model.
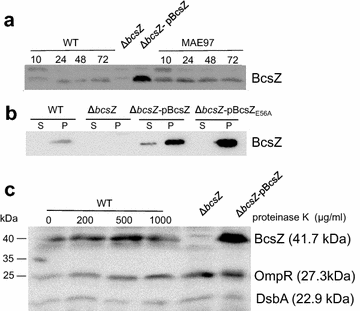
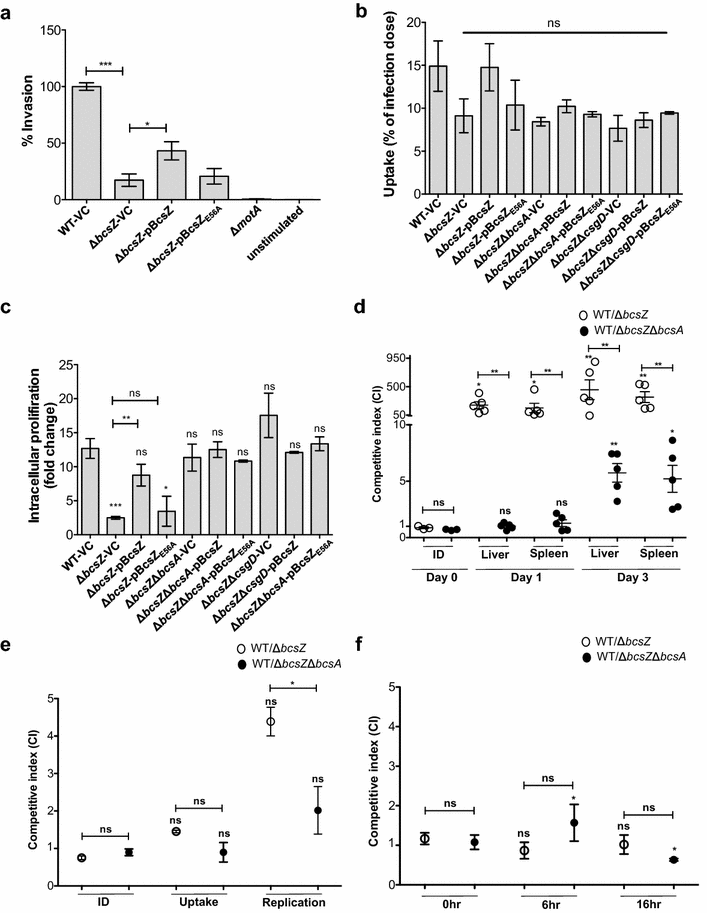
Result: In S. Typhimurium, cellulase BcsZ with a putative periplasmic location negatively regulates cellulose biosynthesis. Moreover, as assessed with a non-polar mutant, BcsZ affects cellulose-associated phenotypes such as the rdar biofilm morphotype, cell clumping, biofilm formation, pellicle formation and flagella-dependent motility. Strikingly, although upregulation of cellulose biosynthesis was not observed on agar plate medium at 37 °C, BcsZ is required for efficient pathogen-host interaction. Key virulence phenotypes of S. Typhimurium such as invasion of epithelial cells and proliferation in macrophages were positively regulated by BcsZ. Further on, a bcsZ mutant was outcompeted by the wild type in organ colonization in the murine typhoid fever infection model. Selected phenotypes were relieved upon deletion of the cellulose synthase BcsA and/or the central biofilm activator CsgD.
Conclusion: Although the protein scaffold has an additional physiological role, our findings indicate that the catalytic activity of BcsZ effectively downregulates CsgD activated cellulose biosynthesis. Repression of cellulose production by BcsZ subsequently enables Salmonella to efficiently colonize the host.
Keywords: BcsZ; Biofilm; Cellulase; Cellulose; CsgD; Salmonella.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

