Rapid detection of pathological mutations and deletions of the haemoglobin beta gene (HBB) by High Resolution Melting (HRM) analysis and Gene Ratio Analysis Copy Enumeration PCR (GRACE-PCR)
- PMID: 27756326
- PMCID: PMC5070150
- DOI: 10.1186/s12881-016-0334-y
Rapid detection of pathological mutations and deletions of the haemoglobin beta gene (HBB) by High Resolution Melting (HRM) analysis and Gene Ratio Analysis Copy Enumeration PCR (GRACE-PCR)
Abstract
Objectives: Inherited disorders of haemoglobin are the world's most common genetic diseases, resulting in significant morbidity and mortality. The large number of mutations associated with the haemoglobin beta gene (HBB) makes gene scanning by High Resolution Melting (HRM) PCR an attractive diagnostic approach. However, existing HRM-PCR assays are not able to detect all common point mutations and have only a very limited ability to detect larger gene rearrangements. The aim of the current study was to develop a HBB assay, which can be used as a screening test in highly heterogeneous populations, for detection of both point mutations and larger gene rearrangements.
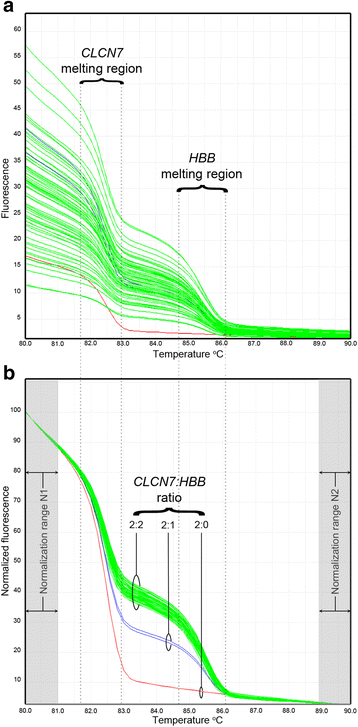
Methods: The assay is based on a combination of conventional HRM-PCR and a novel Gene Ratio Analysis Copy Enumeration (GRACE) PCR method. HRM-PCR was extensively optimised, which included the use of an unlabelled probe and incorporation of universal bases into primers to prevent interference from common non-pathological polymorphisms. GRACE-PCR was employed to determine HBB gene copy numbers relative to a reference gene using melt curve analysis to detect rearrangements in the HBB gene. The performance of the assay was evaluated by analysing 410 samples.
Results: A total of 44 distinct pathological genotypes were detected. In comparison with reference methods, the assay has a sensitivity of 100 % and a specificity of 98 %.
Conclusion: We have developed an assay that detects both point mutations and larger rearrangements of the HBB gene. This assay is quick, sensitive, specific and cost effective making it suitable as an initial screening test that can be used for highly heterogeneous cohorts.
Keywords: Beta thalassaemia; Copy number determination; GRACE-PCR; Gene quantification; HRM.
Figures



References
-
- Thein S. Genetic modifiers of β-thalassemia. Haematologica. 2005;90:649–660. - PubMed
-
- Hbvar. Database of Human Hemoglobin Variants and Thalassemia Mutations. http://globin.bx.psu.edu/hbvar/menu.html. Accessed 1 Sept 2015.
-
- Theodoridou S, Alemayehou M, Prappas N, Karakasidou O, Aletra V, Plata E, Tsaftaridis P, Karababa P, Boussiou M, Sinopoulou K, Hatzi A, Voskaridou E, Loutradi A, Manitsa A. Carrier screening and prenatal diagnosis of hemoglobinopathies. A study of indigenous and immigrant couples in northern Greece, Over the last 5 years. Hemoglobin. 2008;32:434–439. doi: 10.1080/03630260802341745. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

