Prediction of chemo-response in serous ovarian cancer
- PMID: 27756408
- PMCID: PMC5070116
- DOI: 10.1186/s12943-016-0548-9
Prediction of chemo-response in serous ovarian cancer
Abstract
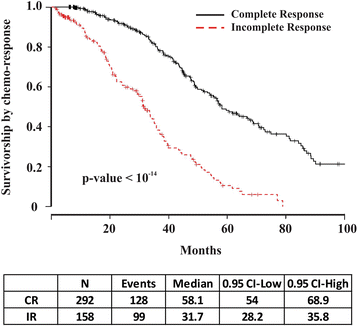
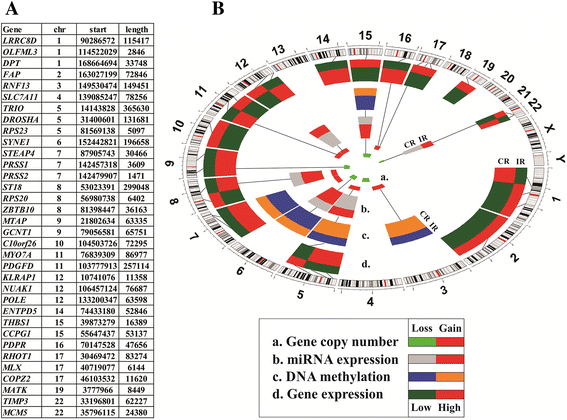
Background: Nearly one-third of serous ovarian cancer (OVCA) patients will not respond to initial treatment with surgery and chemotherapy and die within one year of diagnosis. If patients who are unlikely to respond to current standard therapy can be identified up front, enhanced tumor analyses and treatment regimens could potentially be offered. Using the Cancer Genome Atlas (TCGA) serous OVCA database, we previously identified a robust molecular signature of 422-genes associated with chemo-response. Our objective was to test whether this signature is an accurate and sensitive predictor of chemo-response in serous OVCA.
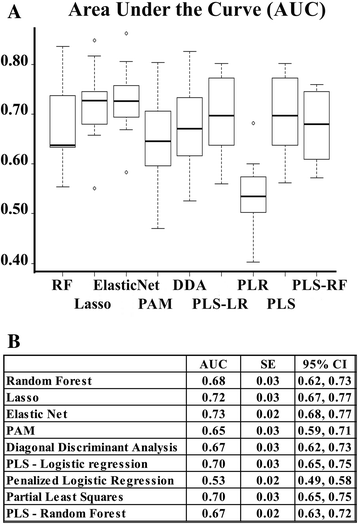
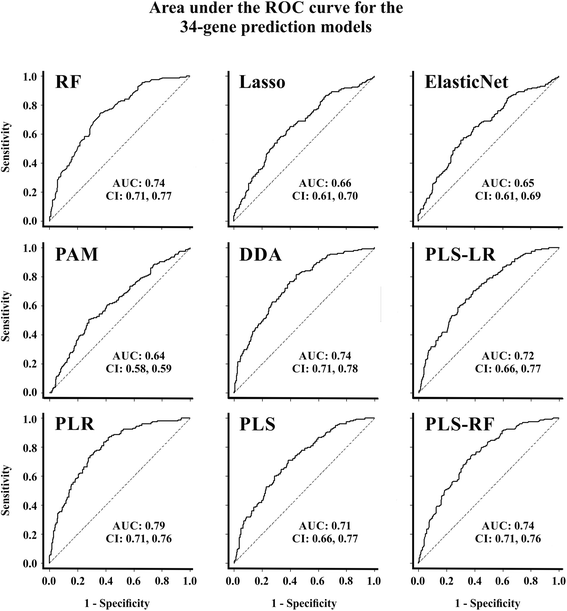
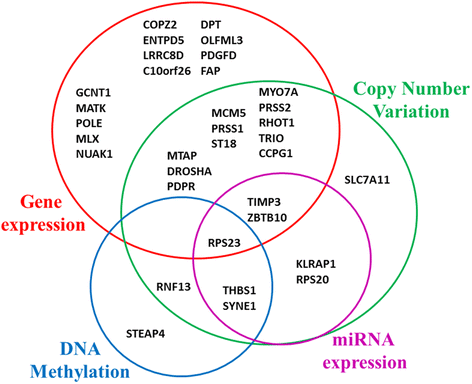
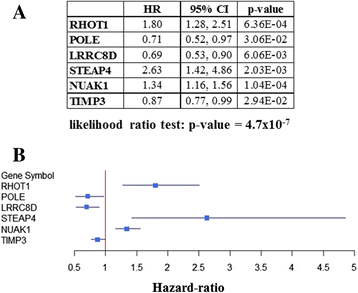
Methods: We first constructed prediction models to predict chemo-response using our previously described 422-gene signature that was associated with response to treatment in serous OVCA. Performance of all prediction models were measured with area under the curves (AUCs, a measure of the model's accuracy) and their respective confidence intervals (CIs). To optimize the prediction process, we determined which elements of the signature most contributed to chemo-response prediction. All prediction models were replicated and validated using six publicly available independent gene expression datasets.
Results: The 422-gene signature prediction models predicted chemo-response with AUCs of ~70 %. Optimization of prediction models identified the 34 most important genes in chemo-response prediction. These 34-gene models had improved performance, with AUCs approaching 80 %. Both 422-gene and 34-gene prediction models were replicated and validated in six independent datasets.
Conclusions: These prediction models serve as the foundation for the future development and implementation of a diagnostic tool to predict response to chemotherapy for serous OVCA patients.
Keywords: Chemo-response; Data integration; Individualized treatment; Ovarian cancer; Prediction model.
Figures
References
-
- American Cancer Society . Cancer Facts & Figures 2014. Atlanta: American Cancer Society; 2014.
-
- Cannistra SA. Cancer of the ovary. N Engl J Med. 2004;351:2519–9. - PubMed
-
- Johnson SW, Laub PB, Beesley JS, Ozols RF, Hamilton TC. Increased platinum-DNA damage tolerance is associated with cisplatin resistance and cross-resistance to various chemotherapeutic agents in unrelated human ovarian cancer cell lines. Cancer Res. 1997;57:850–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

