Dynamic Interplay between Nucleoid Segregation and Genome Integrity in Chlamydomonas Chloroplasts
- PMID: 27756821
- PMCID: PMC5129732
- DOI: 10.1104/pp.16.01533
Dynamic Interplay between Nucleoid Segregation and Genome Integrity in Chlamydomonas Chloroplasts
Abstract
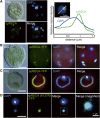
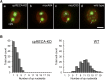
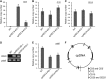
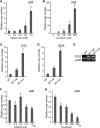
The chloroplast (cp) genome is organized as nucleoids that are dispersed throughout the cp stroma. Previously, a cp homolog of bacterial recombinase RecA (cpRECA) was shown to be involved in the maintenance of cp genome integrity by repairing damaged chloroplast DNA and by suppressing aberrant recombination between short dispersed repeats in the moss Physcomitrella patens Here, overexpression and knockdown analysis of cpRECA in the green alga Chlamydomonas reinhardtii revealed that cpRECA was involved in cp nucleoid dynamics as well as having a role in maintaining cp genome integrity. Overexpression of cpRECA tagged with yellow fluorescent protein or hemagglutinin resulted in the formation of giant filamentous structures that colocalized exclusively to chloroplast DNA and cpRECA localized to cp nucleoids in a heterogenous manner. Knockdown of cpRECA led to a significant reduction in cp nucleoid number that was accompanied by nucleoid enlargement. This phenotype resembled those of gyrase inhibitor-treated cells and monokaryotic chloroplast mutant cells and suggested that cpRECA was involved in organizing cp nucleoid dynamics. The cp genome also was destabilized by induced recombination between short dispersed repeats in cpRECA-knockdown cells and gyrase inhibitor-treated cells. Taken together, these results suggest that cpRECA and gyrase are both involved in nucleoid dynamics and the maintenance of genome integrity and that the mechanisms underlying these processes may be intimately related in C. reinhardtii cps.
© 2016 American Society of Plant Biologists. All Rights Reserved.
Figures

Similar articles
-
RecA maintains the integrity of chloroplast DNA molecules in Arabidopsis.J Exp Bot. 2010 Jun;61(10):2575-88. doi: 10.1093/jxb/erq088. Epub 2010 Apr 20. J Exp Bot. 2010. PMID: 20406785 Free PMC article.
-
RECA plays a dual role in the maintenance of chloroplast genome stability in Physcomitrella patens.Plant J. 2015 Nov;84(3):516-26. doi: 10.1111/tpj.13017. Epub 2015 Oct 15. Plant J. 2015. PMID: 26340426
-
The Chlamydomonas chloroplast HLP protein is required for nucleoid organization and genome maintenance.Mol Plant. 2009 Nov;2(6):1223-32. doi: 10.1093/mp/ssp083. Epub 2009 Oct 29. Mol Plant. 2009. PMID: 19995727
-
Chlamydomonas reinhardtii as the photosynthetic yeast.Annu Rev Genet. 1995;29:209-30. doi: 10.1146/annurev.ge.29.120195.001233. Annu Rev Genet. 1995. PMID: 8825474 Review.
-
Tools and techniques for chloroplast transformation of Chlamydomonas.Adv Exp Med Biol. 2007;616:34-45. doi: 10.1007/978-0-387-75532-8_4. Adv Exp Med Biol. 2007. PMID: 18161489 Review.
Cited by
-
Holliday Junction Resolvase MOC1 Maintains Plastid and Mitochondrial Genome Integrity in Algae and Bryophytes.Plant Physiol. 2020 Dec;184(4):1870-1883. doi: 10.1104/pp.20.00763. Epub 2020 Sep 25. Plant Physiol. 2020. PMID: 32978278 Free PMC article.
-
Chloroplast nucleoids as a transformable network revealed by live imaging with a microfluidic device.Commun Biol. 2018 May 17;1:47. doi: 10.1038/s42003-018-0055-1. eCollection 2018. Commun Biol. 2018. PMID: 30271930 Free PMC article.
-
Factors Affecting Organelle Genome Stability in Physcomitrella patens.Plants (Basel). 2020 Jan 23;9(2):145. doi: 10.3390/plants9020145. Plants (Basel). 2020. PMID: 31979236 Free PMC article. Review.
References
-
- Bungard RA. (2004) Photosynthetic evolution in parasitic plants: insight from the chloroplast genome. BioEssays 26: 235–247 - PubMed
-
- Chen CR, Malik M, Snyder M, Drlica K (1996) DNA gyrase and topoisomerase IV on the bacterial chromosome: quinolone-induced DNA cleavage. J Mol Biol 258: 627–637 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

