A novel gene cluster allows preferential utilization of fucosylated milk oligosaccharides in Bifidobacterium longum subsp. longum SC596
- PMID: 27756904
- PMCID: PMC5069460
- DOI: 10.1038/srep35045
A novel gene cluster allows preferential utilization of fucosylated milk oligosaccharides in Bifidobacterium longum subsp. longum SC596
Abstract
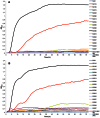
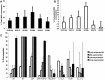
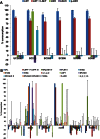
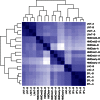
The infant intestinal microbiota is often colonized by two subspecies of Bifidobacterium longum: subsp. infantis (B. infantis) and subsp. longum (B. longum). Competitive growth of B. infantis in the neonate intestine has been linked to the utilization of human milk oligosaccharides (HMO). However, little is known how B. longum consumes HMO. In this study, infant-borne B. longum strains exhibited varying HMO growth phenotypes. While all strains efficiently utilized lacto-N-tetraose, certain strains additionally metabolized fucosylated HMO. B. longum SC596 grew vigorously on HMO, and glycoprofiling revealed a preference for consumption of fucosylated HMO. Transcriptomes of SC596 during early-stage growth on HMO were more similar to growth on fucosyllactose, transiting later to a pattern similar to growth on neutral HMO. B. longum SC596 contains a novel gene cluster devoted to the utilization of fucosylated HMO, including genes for import of fucosylated molecules, fucose metabolism and two α-fucosidases. This cluster showed a modular induction during early growth on HMO and fucosyllactose. This work clarifies the genomic and physiological variation of infant-borne B. longum to HMO consumption, which resembles B. infantis. The capability to preferentially consume fucosylated HMO suggests a competitive advantage for these unique B. longum strains in the breast-fed infant gut.
Conflict of interest statement
J.B.G., C.B.L. and D.A.M., are co-founders of Evolve Biosystems, Inc., a company focused on diet-based manipulation of the gut microbiota.
Figures


Similar articles
-
Fucosyllactose and L-fucose utilization of infant Bifidobacterium longum and Bifidobacterium kashiwanohense.BMC Microbiol. 2016 Oct 26;16(1):248. doi: 10.1186/s12866-016-0867-4. BMC Microbiol. 2016. PMID: 27782805 Free PMC article.
-
Variation in consumption of human milk oligosaccharides by infant gut-associated strains of Bifidobacterium breve.Appl Environ Microbiol. 2013 Oct;79(19):6040-9. doi: 10.1128/AEM.01843-13. Epub 2013 Jul 26. Appl Environ Microbiol. 2013. PMID: 23892749 Free PMC article.
-
Broad conservation of milk utilization genes in Bifidobacterium longum subsp. infantis as revealed by comparative genomic hybridization.Appl Environ Microbiol. 2010 Nov;76(22):7373-81. doi: 10.1128/AEM.00675-10. Epub 2010 Aug 27. Appl Environ Microbiol. 2010. PMID: 20802066 Free PMC article.
-
Human milk oligosaccharides combine with Bifidobacterium longum to form the "golden shield" of the infant intestine: metabolic strategies, health effects, and mechanisms of action.Gut Microbes. 2024 Jan-Dec;16(1):2430418. doi: 10.1080/19490976.2024.2430418. Epub 2024 Nov 21. Gut Microbes. 2024. PMID: 39572856 Free PMC article. Review.
-
Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides.Trends Microbiol. 2010 Jul;18(7):298-307. doi: 10.1016/j.tim.2010.03.008. Epub 2010 Apr 19. Trends Microbiol. 2010. PMID: 20409714 Free PMC article. Review.
Cited by
-
Gold standard for nutrition: a review of human milk oligosaccharide and its effects on infant gut microbiota.Microb Cell Fact. 2021 May 28;20(1):108. doi: 10.1186/s12934-021-01599-y. Microb Cell Fact. 2021. PMID: 34049536 Free PMC article. Review.
-
Bifidobacterium longum and microbiome maturation modify a nutrient intervention for stunting in Zimbabwean infants.EBioMedicine. 2024 Oct;108:105362. doi: 10.1016/j.ebiom.2024.105362. Epub 2024 Sep 27. EBioMedicine. 2024. PMID: 39341154 Free PMC article.
-
Mechanism of 2'-fucosyllactose degradation by human-associated Akkermansia.J Bacteriol. 2024 Feb 22;206(2):e0033423. doi: 10.1128/jb.00334-23. Epub 2024 Feb 1. J Bacteriol. 2024. PMID: 38299857 Free PMC article.
-
[A review on the relationship between breast milk nutrients and brain development in preterm infants].Zhongguo Dang Dai Er Ke Za Zhi. 2019 Jun;21(6):607-612. doi: 10.7499/j.issn.1008-8830.2019.06.020. Zhongguo Dang Dai Er Ke Za Zhi. 2019. PMID: 31208518 Free PMC article. Review. Chinese.
-
Identification of plasmalogens in Bifidobacterium longum, but not in Bifidobacterium animalis.Sci Rep. 2020 Jan 16;10(1):427. doi: 10.1038/s41598-019-57309-7. Sci Rep. 2020. PMID: 31949186 Free PMC article.
References
-
- Sekirov I., Russell S. L., Antunes L. C. M. & Finlay B. B. Gut Microbiota in Health and Disease. Physiological Reviews 90, 859–904 (2010). - PubMed
-
- Zeissig S. & Blumberg R. S. Life at the beginning: perturbation of the microbiota by antibiotics in early life and its role in health and disease. Nat Immunol 15, 307–310 (2014). - PubMed
-
- Scholtens P. A., Oozeer R., Martin R., Amor K. B. & Knol J. The early settlers: intestinal microbiology in early life. Annu Rev Food Sci Technol 3, 425–447 (2012). - PubMed
-
- Kalliomaki M., Collado M. C., Salminen S. & Isolauri E. Early differences in fecal microbiota composition in children may predict overweight. Am J Clin Nutr 87, 534–538 (2008). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

