A Novel Fucose-binding Lectin from Photorhabdus luminescens (PLL) with an Unusual Heptabladed β-Propeller Tetrameric Structure
- PMID: 27758853
- PMCID: PMC5122772
- DOI: 10.1074/jbc.M115.693473
A Novel Fucose-binding Lectin from Photorhabdus luminescens (PLL) with an Unusual Heptabladed β-Propeller Tetrameric Structure
Abstract
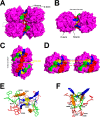
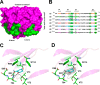
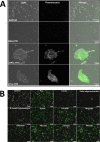
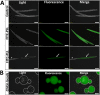
Photorhabdus luminescens is known for its symbiosis with the entomopathogenic nematode Heterorhabditis bacteriophora and its pathogenicity toward insect larvae. A hypothetical protein from P. luminescens was identified, purified from the native source, and characterized as an l-fucose-binding lectin, named P. luminescens lectin (PLL). Glycan array and biochemical characterization data revealed PLL to be specific toward l-fucose and the disaccharide glycan 3,6-O-Me2-Glcβ1-4(2,3-O-Me2)Rhaα-O-(p-C6H4)-OCH2CH2NH2 PLL was discovered to be a homotetramer with an intersubunit disulfide bridge. The crystal structures of native and recombinant PLL revealed a seven-bladed β-propeller fold creating seven putative fucose-binding sites per monomer. The crystal structure of the recombinant PLL·l-fucose complex confirmed that at least three sites were fucose-binding. Moreover, the crystal structures indicated that some of the other sites are masked either by the tetrameric nature of the lectin or by incorporation of the C terminus of the lectin into one of these sites. PLL exhibited an ability to bind to insect hemocytes and the cuticular surface of a nematode, H. bacteriophora.
Keywords: Galleria mellonella; Photorhabdus luminescens; bacterial pathogenesis; crystal structure; hemocytes from insect larvae; host/pathogen interaction; lectin; structural biology.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


Similar articles
-
Lectin PLL3, a Novel Monomeric Member of the Seven-Bladed β-Propeller Lectin Family.Molecules. 2019 Dec 11;24(24):4540. doi: 10.3390/molecules24244540. Molecules. 2019. PMID: 31835851 Free PMC article.
-
Characterization of novel bangle lectin from Photorhabdus asymbiotica with dual sugar-binding specificity and its effect on host immunity.PLoS Pathog. 2017 Aug 14;13(8):e1006564. doi: 10.1371/journal.ppat.1006564. eCollection 2017 Aug. PLoS Pathog. 2017. PMID: 28806750 Free PMC article.
-
The insight into the biology of five homologous lectins produced by the entomopathogenic bacterium and nematode symbiont Photorhabdus laumondii.Glycobiology. 2025 Jun 2;35(7):cwaf033. doi: 10.1093/glycob/cwaf033. Glycobiology. 2025. PMID: 40459235 Free PMC article.
-
Regulation of Phenotypic Switching and Heterogeneity in Photorhabdus luminescens Cell Populations.J Mol Biol. 2019 Nov 22;431(23):4559-4568. doi: 10.1016/j.jmb.2019.04.015. Epub 2019 Apr 22. J Mol Biol. 2019. PMID: 31022406 Review.
-
F-Type Lectins: A Highly Diversified Family of Fucose-Binding Proteins with a Unique Sequence Motif and Structural Fold, Involved in Self/Non-Self-Recognition.Front Immunol. 2017 Nov 29;8:1648. doi: 10.3389/fimmu.2017.01648. eCollection 2017. Front Immunol. 2017. PMID: 29238345 Free PMC article. Review.
Cited by
-
Fucose-binding lectins: purification, characterization and potential biomedical applications.Mol Biol Rep. 2023 Dec;50(12):10589-10603. doi: 10.1007/s11033-023-08896-2. Epub 2023 Nov 7. Mol Biol Rep. 2023. PMID: 37934371 Review.
-
Photorhabdus luminescens lectin A (PllA): A new probe for detecting α-galactoside-terminating glycoconjugates.J Biol Chem. 2017 Dec 1;292(48):19935-19951. doi: 10.1074/jbc.M117.812792. Epub 2017 Sep 28. J Biol Chem. 2017. PMID: 28972138 Free PMC article.
-
Overview of the Structure⁻Function Relationships of Mannose-Specific Lectins from Plants, Algae and Fungi.Int J Mol Sci. 2019 Jan 10;20(2):254. doi: 10.3390/ijms20020254. Int J Mol Sci. 2019. PMID: 30634645 Free PMC article. Review.
-
Lectin PLL3, a Novel Monomeric Member of the Seven-Bladed β-Propeller Lectin Family.Molecules. 2019 Dec 11;24(24):4540. doi: 10.3390/molecules24244540. Molecules. 2019. PMID: 31835851 Free PMC article.
-
Fucose as a nutrient ligand for Dikarya and a building block of early diverging lineages.IMA Fungus. 2023 Sep 5;14(1):17. doi: 10.1186/s43008-023-00123-8. IMA Fungus. 2023. PMID: 37670396 Free PMC article.
References
-
- Joyce S. A., Watson R. J., and Clarke D. J. (2006) The regulation of pathogenicity and mutualism in Photorhabdus. Curr. Opin. Microbiol. 9, 127–132 - PubMed
-
- Forst S., Dowds B., Boemare N., and Stackebrandt E. (1997) Xenorhabdus and Photorhabdus spp.: bugs that kill bugs. Annu. Rev. Microbiol. 51, 47–72 - PubMed
-
- Duchaud E., Rusniok C., Frangeul L., Buchrieser C., Givaudan A., Taourit S., Bocs S., Boursaux-Eude C., Chandler M., Charles J. F., Dassa E., Derose R., Derzelle S., Freyssinet G., Gaudriault S., et al. (2003) The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 21, 1307–13 - PubMed
-
- Williamson V. M., and Kaya H. K. (2003) Sequence of a symbiont. Nat. Biotechnol. 21, 1294–1295 - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

