Oncogenic Functions of Gli1 in Pancreatic Adenocarcinoma Are Supported by Its PRMT1-Mediated Methylation
- PMID: 27758883
- PMCID: PMC5135656
- DOI: 10.1158/0008-5472.CAN-16-0715
Oncogenic Functions of Gli1 in Pancreatic Adenocarcinoma Are Supported by Its PRMT1-Mediated Methylation
Abstract
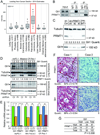
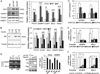
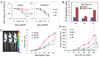
The oncogenic transcription factor Gli1 is a critical effector in the Hedgehog (Hh) pathway, which is necessary for the development and progression of pancreatic ductal adenocarcinoma (PDAC). Although TGFβ and K-Ras are known regulators of Gli1 gene transcription in this setting, it is not understood how Gli1 functional activity is regulated. Here, we report the identification of Gli1 as a substrate for the protein arginine N-methyltransferase PRMT1 in PDAC. We found that PRMT1 methylates Gli1 at R597, promoting its transcriptional activity by enhancing the binding of Gli1 to its target gene promoters. Interruption of Gli1 methylation attenuates oncogenic functions of Gli1 and sensitizes PDAC cells to gemcitabine treatment. In human PDAC specimens, the levels of both total Gli1 and methylated Gli1 were correlated positively with PRMT1 protein levels. Notably, PRMT1 regulated Gli1 independently of the canonical Hh pathway as well as the TGFβ/Kras-mediated noncanonical Hh pathway, thereby signifying a novel regulatory mechanism for Gli1 transcriptional activity. Taken together, our results identified a new posttranslational modification of Gli1 that underlies its pivotal oncogenic functions in PDAC. Cancer Res; 76(23); 7049-58. ©2016 AACR.
©2016 American Association for Cancer Research.
Conflict of interest statement
The authors have declared that no conflict of interest exists.
Figures


Similar articles
-
Protein arginine methyltransferase 1 interacts with Gli1 and regulates its transcriptional activity.Tumour Biol. 2016 Jul;37(7):9071-6. doi: 10.1007/s13277-015-4754-7. Epub 2016 Jan 13. Tumour Biol. 2016. PMID: 26762411
-
GLI1, a master regulator of the hallmark of pancreatic cancer.Pathol Int. 2016 Dec;66(12):653-660. doi: 10.1111/pin.12476. Epub 2016 Nov 8. Pathol Int. 2016. PMID: 27862693 Review.
-
SHh-Gli1 signaling pathway promotes cell survival by mediating baculoviral IAP repeat-containing 3 (BIRC3) gene in pancreatic cancer cells.Tumour Biol. 2016 Jul;37(7):9943-50. doi: 10.1007/s13277-016-4898-0. Epub 2016 Jan 27. Tumour Biol. 2016. PMID: 26815504
-
GLI1 interferes with the DNA mismatch repair system in pancreatic cancer through BHLHE41-mediated suppression of MLH1.Cancer Res. 2013 Dec 15;73(24):7313-23. doi: 10.1158/0008-5472.CAN-13-2008. Epub 2013 Oct 28. Cancer Res. 2013. PMID: 24165159
-
Targeting hedgehog signaling in pancreatic ductal adenocarcinoma.Pharmacol Ther. 2022 Aug;236:108107. doi: 10.1016/j.pharmthera.2022.108107. Epub 2022 Jan 7. Pharmacol Ther. 2022. PMID: 34999181 Review.
Cited by
-
Role and inhibition of GLI1 protein in cancer.Lung Cancer (Auckl). 2018 Mar 27;9:35-43. doi: 10.2147/LCTT.S124483. eCollection 2018. Lung Cancer (Auckl). 2018. PMID: 29628779 Free PMC article. Review.
-
PRMT1-dependent regulation of RNA metabolism and DNA damage response sustains pancreatic ductal adenocarcinoma.Nat Commun. 2021 Jul 30;12(1):4626. doi: 10.1038/s41467-021-24798-y. Nat Commun. 2021. PMID: 34330913 Free PMC article.
-
Therapeutic targeting potential of the protein lysine and arginine methyltransferases to reverse cancer chemoresistance.Front Mol Biosci. 2024 Dec 5;11:1455415. doi: 10.3389/fmolb.2024.1455415. eCollection 2024. Front Mol Biosci. 2024. PMID: 39703687 Free PMC article. Review.
-
Fine-Tuning of GLI Activity through Arginine Methylation: Its Mechanisms and Function.Cells. 2020 Aug 26;9(9):1973. doi: 10.3390/cells9091973. Cells. 2020. PMID: 32859041 Free PMC article. Review.
-
PRMT1 promotes pancreatic cancer growth and predicts poor prognosis.Cell Oncol (Dordr). 2020 Feb;43(1):51-62. doi: 10.1007/s13402-019-00435-1. Epub 2019 Sep 13. Cell Oncol (Dordr). 2020. PMID: 31520395
References
-
- Mao J, Maye P, Kogerman P, Tejedor FJ, Toftgard R, Xie W, et al. Regulation of Gli1 transcriptional activity in the nucleus by Dyrk1. The Journal of biological chemistry. 2002;277(38):35156–35161. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

