Whole-exome sequencing identifies novel variants in PNPT1 causing oxidative phosphorylation defects and severe multisystem disease
- PMID: 27759031
- PMCID: PMC5159763
- DOI: 10.1038/ejhg.2016.128
Whole-exome sequencing identifies novel variants in PNPT1 causing oxidative phosphorylation defects and severe multisystem disease
Abstract
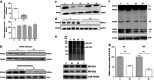
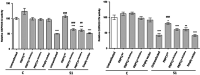
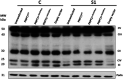
Recent advances in next-generation sequencing strategies have led to the discovery of many novel disease genes. We describe here a non-consanguineous family with two affected boys presenting with early onset of severe axonal neuropathy, optic atrophy, intellectual disability, auditory neuropathy and chronic respiratory and gut disturbances. Whole-exome sequencing (WES) was performed on all family members and we identified compound heterozygous variants (c.[760C>A];[1528G>C];p.[(Gln254Lys);(Ala510Pro)] in the polyribonucleotide nucleotidyltransferase 1 (PNPT1) gene in both affected individuals. PNPT1 encodes the polynucleotide phosphorylase (PNPase) protein, which is involved in the transport of small RNAs into the mitochondria. These RNAs are involved in the mitochondrial translation machinery, responsible for the synthesis of mitochondrially encoded subunits of the oxidative phosphorylation (OXPHOS) complexes. Both PNPT1 variants are within highly conserved regions and predicted to be damaging. These variants resulted in quaternary defects in the PNPase protein and a clear reduction in protein and mRNA expression of PNPT1 in patient fibroblasts compared with control cells. Protein analysis of the OXPHOS complexes showed a significant reduction in complex I (CI), complex III (CIII) and complex IV (CIV). Enzyme activity of CI and CIV was clearly reduced in patient fibroblasts compared with controls along with a 33% reduction in total mitochondrial protein synthesis. In vitro rescue experiments, using exogenous expression of wild-type PNPT1 in patient fibroblasts, ameliorated the deficiencies in the OXPHOS complex protein expression, supporting the likely pathogenicity of these variants and the importance of WES in efficiently identifying rare genetic disease genes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Menezes MJ, Riley LG, Christodoulou J: Mitochondrial respiratory chain disorders in childhood: insights into diagnosis and management in the new era of genomic medicine. Biochim Biophys Acta 2014; 1840: 1368–1379. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

