Crystal structure of human aldehyde dehydrogenase 1A3 complexed with NAD+ and retinoic acid
- PMID: 27759097
- PMCID: PMC5069622
- DOI: 10.1038/srep35710
Crystal structure of human aldehyde dehydrogenase 1A3 complexed with NAD+ and retinoic acid
Abstract
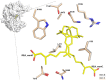
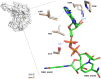
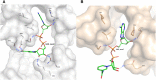
The aldehyde dehydrogenase family 1 member A3 (ALDH1A3) catalyzes the oxidation of retinal to the pleiotropic factor retinoic acid using NAD+. The level of ALDHs enzymatic activity has been used as a cancer stem cell marker and seems to correlate with tumour aggressiveness. Elevated ALDH1A3 expression in mesenchymal glioma stem cells highlights the potential of this isozyme as a prognosis marker and drug target. Here we report the first crystal structure of human ALDH1A3 complexed with NAD+ and the product all-trans retinoic acid (REA). The tetrameric ALDH1A3 folds into a three domain-based architecture highly conserved along the ALDHs family. The structural analysis revealed two different and coupled conformations for NAD+ and REA that we propose to represent two snapshots along the catalytic cycle. Indeed, the isoprenic moiety of REA points either toward the active site cysteine, or moves away adopting the product release conformation. Although ALDH1A3 shares high sequence identity with other members of the ALDH1A family, our structural analysis revealed few peculiar residues in the 1A3 isozyme active site. Our data provide information into the ALDH1As catalytic process and can be used for the structure-based design of selective inhibitors of potential medical interest.
Figures




References
-
- Vasiliou V., Pappa A. & Petersen D. R. Role of aldehyde dehydrogenases in endogenous and xenobiotic metabolism. Chem Biol Interact. 129, 1–19 (2000). - PubMed
-
- Vasiliou V., Pappa A. & Estey T. Role of human aldehyde dehydrogenases in endobiotic and xenobiotic metabolism. Drug Metab Rev. 36, 279–299 (2004). - PubMed
-
- Yokoyama A. et al. Alcohol and aldehyde dehydrogenase gene polymorphisms and oropharyngolaryngeal, esophageal and stomach cancers in Japanese alcoholics. Carcinogenesis 22, 433–439 (2001). - PubMed
-
- Sladek N. E. Human aldehyde dehydrogenases: potential pathological, pharmacological, and toxicological impact. J Biochem Mol Toxicol. 17, 7–23 (2003). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

