Helicase CHD4 is an epigenetic coregulator of PAX3-FOXO1 in alveolar rhabdomyosarcoma
- PMID: 27760049
- PMCID: PMC5096911
- DOI: 10.1172/JCI85057
Helicase CHD4 is an epigenetic coregulator of PAX3-FOXO1 in alveolar rhabdomyosarcoma
Abstract
A vast number of cancer genes are transcription factors that drive tumorigenesis as oncogenic fusion proteins. Although the direct targeting of transcription factors remains challenging, therapies aimed at oncogenic fusion proteins are attractive as potential treatments for cancer. There is particular interest in targeting the oncogenic PAX3-FOXO1 fusion transcription factor, which induces alveolar rhabdomyosarcoma (aRMS), an aggressive cancer of skeletal muscle cells for which patient outcomes remain dismal. In this work, we have defined the interactome of PAX3-FOXO1 and screened 60 candidate interactors using siRNA-mediated depletion to identify candidates that affect fusion protein activity in aRMS cells. We report that chromodomain helicase DNA binding protein 4 (CHD4), an ATP-dependent chromatin remodeler, acts as crucial coregulator of PAX3-FOXO1 activity. CHD4 interacts with PAX3-FOXO1 via short DNA fragments. Together, they bind to regulatory regions of PAX3-FOXO1 target genes. Gene expression analysis suggested that CHD4 coregulatory activity is essential for a subset of PAX3-FOXO1 target genes. Depletion of CHD4 reduced cell viability of fusion-positive but not of fusion-negative RMS in vitro, which resembled loss of PAX3-FOXO1. It also caused specific regression of fusion-positive xenograft tumors in vivo. Therefore, this work identifies CHD4 as an epigenetic coregulator of PAX3-FOXO1 activity, providing rational evidence for CHD4 as a potential therapeutic target in aRMS.
Figures
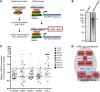


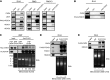
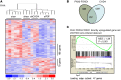

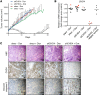
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

