Formin DAAM1 Organizes Actin Filaments in the Cytoplasmic Nodal Actin Network
- PMID: 27760153
- PMCID: PMC5070803
- DOI: 10.1371/journal.pone.0163915
Formin DAAM1 Organizes Actin Filaments in the Cytoplasmic Nodal Actin Network
Abstract
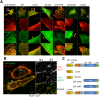
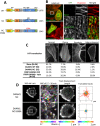
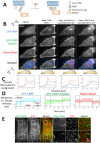
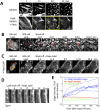
A nodal cytoplasmic actin network underlies actin cytoplasm cohesion in the absence of stress fibers. We previously described such a network that forms upon Latrunculin A (LatA) treatment, in which formin DAAM1 was localized at these nodes. Knock down of DAAM1 reduced the mobility of actin nodes but the nodes remained. Here we have investigated DAAM1 containing nodes after LatA washout. DAAM1 was found to be distributed between the cytoplasm and the plasma membrane. The membrane binding likely occurs through an interaction with lipid rafts, but is not required for F-actin assembly. Interesting the forced interaction of DAAM1 with plasma membrane through a rapamycin-dependent linkage, enhanced F-actin assembly at the cell membrane (compared to the cytoplasm) after the LatA washout. However, immediately after addition of both rapamycin and LatA, the cytoplasmic actin nodes formed transiently, before DAAM1 moved to the membrane. This was consistent with the idea that DAAM1 was initially anchored to cytoplasmic actin nodes. Further, photoactivatable tracking of DAAM1 showed DAAM1 was immobilized at these actin nodes. Thus, we suggest that DAAM1 organizes actin filaments into a nodal complex, and such nodal complexes seed actin network recovery after actin depolymerization.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Analysis of the local organization and dynamics of cellular actin networks.J Cell Biol. 2013 Sep 30;202(7):1057-73. doi: 10.1083/jcb.201210123. J Cell Biol. 2013. PMID: 24081490 Free PMC article.
-
Flightless-I (Fli-I) regulates the actin assembly activity of diaphanous-related formins (DRFs) Daam1 and mDia1 in cooperation with active Rho GTPase.J Biol Chem. 2010 May 21;285(21):16231-8. doi: 10.1074/jbc.M109.079236. Epub 2010 Mar 11. J Biol Chem. 2010. PMID: 20223827 Free PMC article.
-
Daam1 regulates fascin for actin assembly in mouse oocyte meiosis.Cell Cycle. 2017 Jul 18;16(14):1350-1356. doi: 10.1080/15384101.2017.1325045. Epub 2017 Jul 6. Cell Cycle. 2017. PMID: 28682694 Free PMC article.
-
FHOD proteins in actin dynamics--a formin' class of its own.Small GTPases. 2014;5(2):11. doi: 10.4161/21541248.2014.973765. Small GTPases. 2014. PMID: 25483300 Free PMC article. Review.
-
Cytoplasmic zoning in membrane blebs.J Biochem. 2024 Feb 25;175(2):133-140. doi: 10.1093/jb/mvad084. J Biochem. 2024. PMID: 37943501 Review.
Cited by
-
Parent of origin gene expression in a founder population identifies two new candidate imprinted genes at known imprinted regions.PLoS One. 2018 Sep 11;13(9):e0203906. doi: 10.1371/journal.pone.0203906. eCollection 2018. PLoS One. 2018. PMID: 30204804 Free PMC article.
-
Adaptive optics improves multiphoton super-resolution imaging.Nat Methods. 2017 Sep;14(9):869-872. doi: 10.1038/nmeth.4337. Epub 2017 Jun 19. Nat Methods. 2017. PMID: 28628128 Free PMC article.
-
Divergent roles of the Wnt/PCP Formin Daam1 in renal ciliogenesis.PLoS One. 2019 Aug 30;14(8):e0221698. doi: 10.1371/journal.pone.0221698. eCollection 2019. PLoS One. 2019. PMID: 31469868 Free PMC article.
-
Astral architecture can enhance mechanical strength of cytoskeletal networks by modulating percolation thresholds.bioRxiv [Preprint]. 2025 Jun 22:2025.06.17.660175. doi: 10.1101/2025.06.17.660175. bioRxiv. 2025. PMID: 40666881 Free PMC article. Preprint.
-
Differential Expression of microRNAs in Serum of Patients with Chronic Painful Polyneuropathy and Healthy Age-Matched Controls.Biomedicines. 2023 Mar 2;11(3):764. doi: 10.3390/biomedicines11030764. Biomedicines. 2023. PMID: 36979743 Free PMC article.
References
-
- Matsudaira P. Actin crosslinking proteins at the leading edge. Seminars in cell biology. 1994;5(3):165–74. . - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

