Genomic Regions Associated with Feed Efficiency Indicator Traits in an Experimental Nellore Cattle Population
- PMID: 27760167
- PMCID: PMC5070821
- DOI: 10.1371/journal.pone.0164390
Genomic Regions Associated with Feed Efficiency Indicator Traits in an Experimental Nellore Cattle Population
Erratum in
-
Correction: Genomic Regions Associated with Feed Efficiency Indicator Traits in an Experimental Nellore Cattle Population.PLoS One. 2017 Feb 6;12(2):e0171845. doi: 10.1371/journal.pone.0171845. eCollection 2017. PLoS One. 2017. PMID: 28166287 Free PMC article.
Abstract
The objective of this study was to identify genomic regions and metabolic pathways associated with dry matter intake, average daily gain, feed efficiency and residual feed intake in an experimental Nellore cattle population. The high-density SNP chip (Illumina High-Density Bovine BeadChip, 777k) was used to genotype the animals. The SNP markers effects and their variances were estimated using the single-step genome wide association method. The (co)variance components were estimated by Bayesian inference. The chromosome segments that are responsible for more than 1.0% of additive genetic variance were selected to explore and determine possible quantitative trait loci. The bovine genome Map Viewer was used to identify genes. In total, 51 genomic regions were identified for all analyzed traits. The heritability estimated for feed efficiency was low magnitude (0.13±0.06). For average daily gain, dry matter intake and residual feed intake, heritability was moderate to high (0.43±0.05; 0.47±0.05, 0.18±0.05, respectively). A total of 8, 17, 14 and 12 windows that are responsible for more than 1% of the additive genetic variance for dry matter intake, average daily gain, feed efficiency and residual feed intake, respectively, were identified. Candidate genes GOLIM4, RFX6, CACNG7, CACNG6, CAPN8, CAPN2, AKT2, GPRC6A, and GPR45 were associated with feed efficiency traits. It was expected that the response to selection would be higher for residual feed intake than for feed efficiency. Genomic regions harboring possible QTL for feed efficiency indicator traits were identified. Candidate genes identified are involved in energy use, metabolism protein, ion transport, transmembrane transport, the olfactory system, the immune system, secretion and cellular activity. The identification of these regions and their respective candidate genes should contribute to the formation of a genetic basis in Nellore cattle for feed efficiency indicator traits, and these results would support the selection for these traits.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

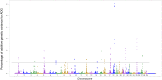
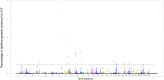

References
-
- Anderson RV, Rasby RJ, Klopfenstein TJ, and Clark RT. An evaluation of production and economic efficiency of two beef systems from calving to slaughter. J Anim Sci. 2005;83: 694–704. - PubMed
-
- Arthur JPF and Herd RM. Residual feed intake in beef cattle. R Bras Zootec. 2008;37: 269–279.
-
- Arthur PF, Archer JA, Johnston DJ, Herd RM, Richardson EC, and Parnell PF. Genetic and phenotypic variance and covariance componentes for feed intake, feed efficiency, and other postweaning traits in Angus cattle. J Anim Sci. 2001;79: 2805–2811. - PubMed
-
- Tedeschi LO, Fox DG, Baker MJ, and Kirschten DP. Identifying differences in feed efficiency among group-fed cattle. J. Anim. Sci. 2006;84: 767–776. - PubMed
-
- Koch RM, Swiger LA, Doyle C, and Gregory KE. Efficiency of feed use in beef cattle. J Anim Sci. 1963;22: 486–494.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

