Elucidation of the Fanconi Anemia Protein Network in Meiosis and Its Function in the Regulation of Histone Modifications
- PMID: 27760317
- PMCID: PMC5095620
- DOI: 10.1016/j.celrep.2016.09.073
Elucidation of the Fanconi Anemia Protein Network in Meiosis and Its Function in the Regulation of Histone Modifications
Abstract
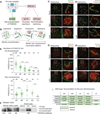
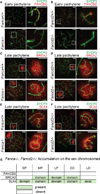
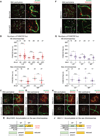
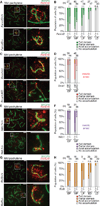
Precise epigenetic regulation of the sex chromosomes is vital for the male germline. Here, we analyze meiosis in eight mouse models deficient for various DNA damage response (DDR) factors, including Fanconi anemia (FA) proteins. We reveal a network of FA and DDR proteins in which FA core factors FANCA, FANCB, and FANCC are essential for FANCD2 foci formation, whereas BRCA1 (FANCS), MDC1, and RNF8 are required for BRCA2 (FANCD1) and SLX4 (FANCP) accumulation on the sex chromosomes during meiosis. In addition, FA proteins modulate distinct histone marks on the sex chromosomes: FA core proteins and FANCD2 regulate H3K9 methylation, while FANCD2 and RNF8 function together to regulate H3K4 methylation independently of FA core proteins. Our data suggest that RNF8 integrates the FA-BRCA pathway. Taken together, our study reveals distinct functions for FA proteins and illuminates the male sex chromosomes as a model to dissect the function of the FA-BRCA pathway.
Keywords: DNA damage response; DNA repair; Fanconi anemia; X chromosome; epigenetics; germ cells; meiosis; meiotic sex chromosome inactivation; sex chromosomes; spermatogenesis.
Copyright © 2016 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Bakker ST, van de Vrugt HJ, Rooimans MA, Oostra AB, Steltenpool J, Delzenne-Goette E, van der Wal A, van der Valk M, Joenje H, te Riele H, de Winter JP. Fancm-deficient mice reveal unique features of Fanconi anemia complementation group M. Hum. Mol. Genet. 2009;18:3484–3495. - PubMed
-
- Baudat F, Manova K, Yuen JP, Jasin M, Keeney S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell. 2000;6:989–998. - PubMed
-
- Bellani MA, Romanienko PJ, Cairatti DA, Camerini-Otero RD. SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm−/− spermatocytes. J. Cell Sci. 2005;118:3233–3245. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

