Closing the loop: 3C versus DNA FISH
- PMID: 27760553
- PMCID: PMC5072311
- DOI: 10.1186/s13059-016-1081-2
Closing the loop: 3C versus DNA FISH
Abstract
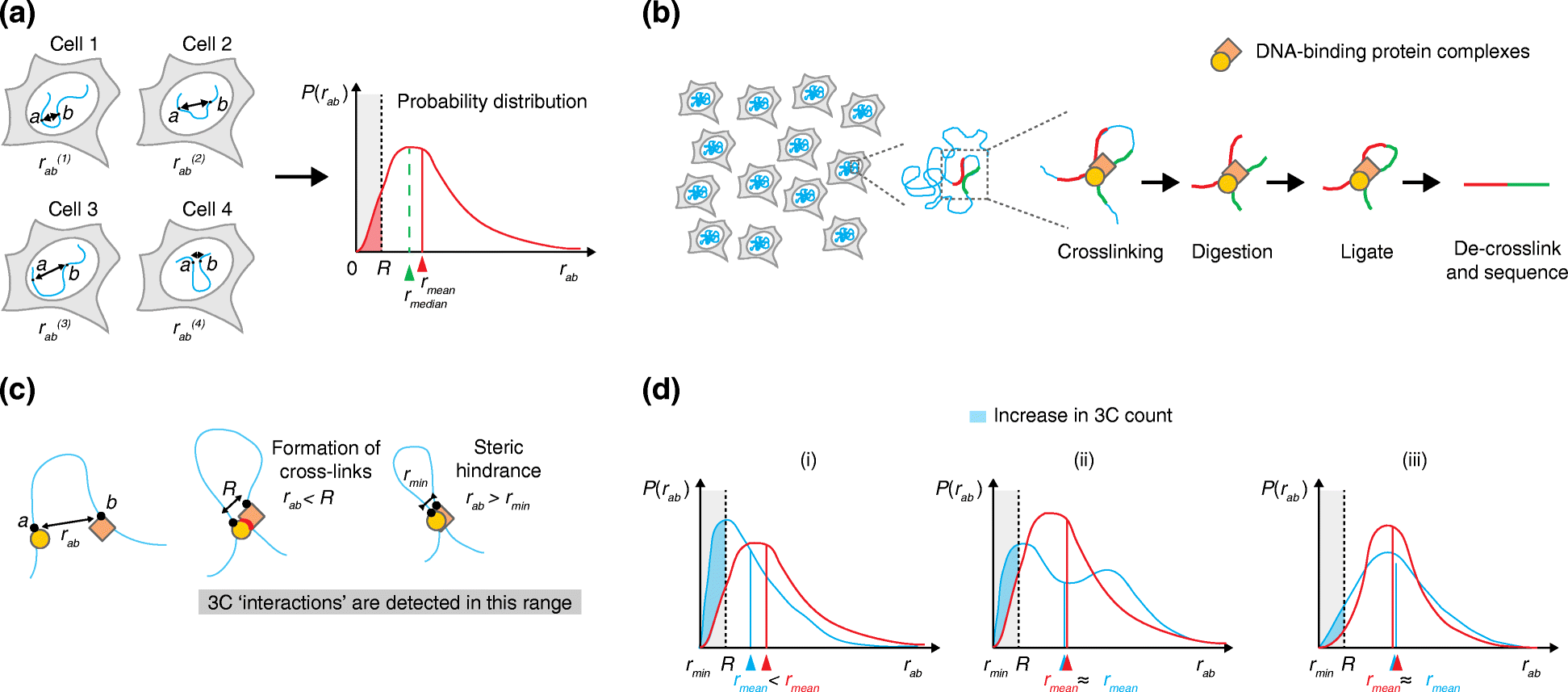
Chromosome conformation capture (3C)-based techniques have revolutionized the field of nuclear organization, partly replacing DNA FISH as the method of choice for studying three-dimensional chromosome architecture. Although DNA FISH is commonly used for confirming 3C-based findings, the two techniques are conceptually and technically different and comparing their results is not trivial. Here, we discuss both 3C-based techniques and DNA FISH approaches to highlight their similarities and differences. We then describe the technical biases that affect each approach, and review the available reports that address their compatibility. Finally, we propose an experimental scheme for comparison of 3C and DNA FISH results.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

