Tiny microbes, enormous impacts: what matters in gut microbiome studies?
- PMID: 27760558
- PMCID: PMC5072314
- DOI: 10.1186/s13059-016-1086-x
Tiny microbes, enormous impacts: what matters in gut microbiome studies?
Abstract
Many factors affect the microbiomes of humans, mice, and other mammals, but substantial challenges remain in determining which of these factors are of practical importance. Considering the relative effect sizes of both biological and technical covariates can help improve study design and the quality of biological conclusions. Care must be taken to avoid technical bias that can lead to incorrect biological conclusions. The presentation of quantitative effect sizes in addition to P values will improve our ability to perform meta-analysis and to evaluate potentially relevant biological effects. A better consideration of effect size and statistical power will lead to more robust biological conclusions in microbiome studies.
Figures
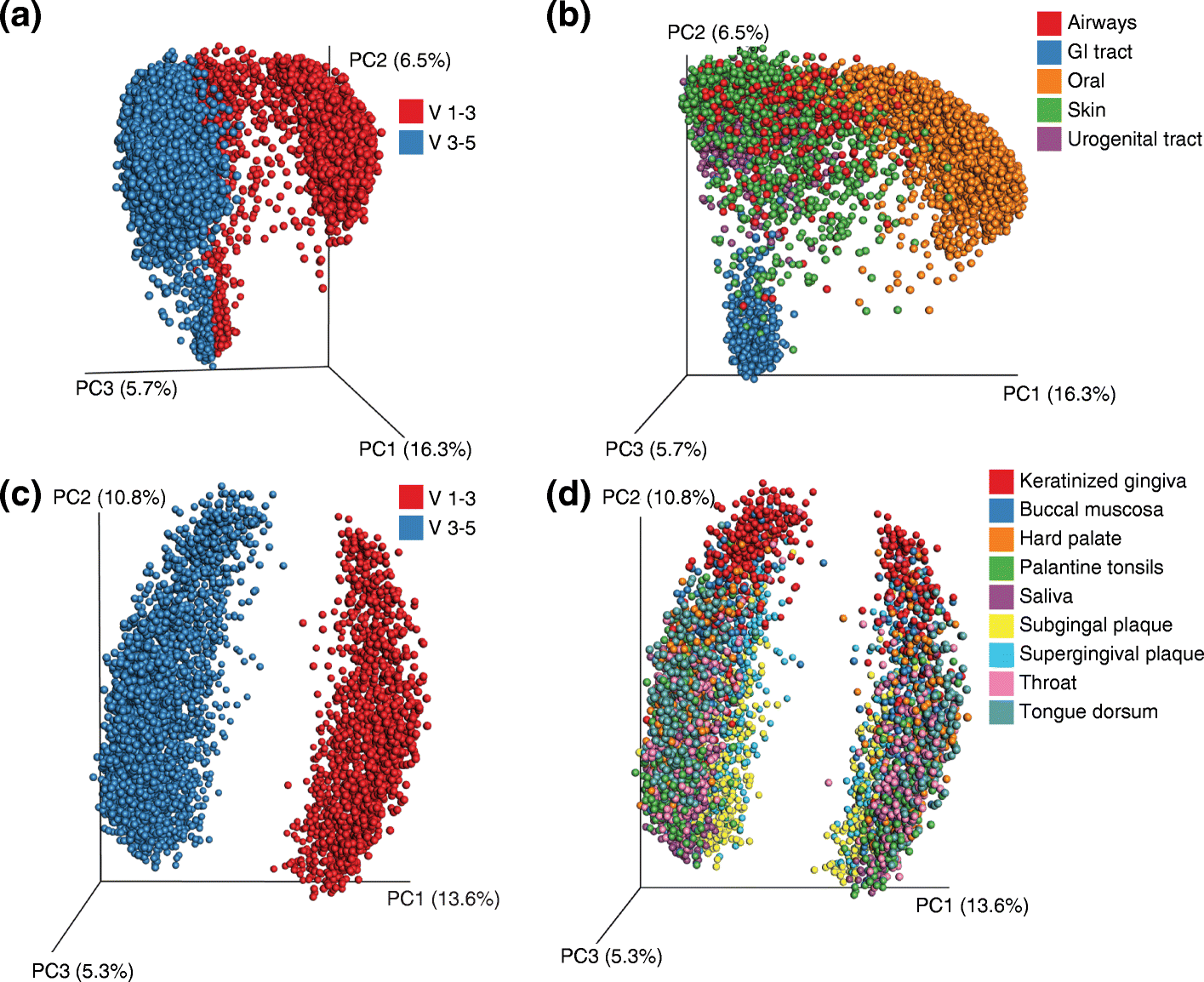
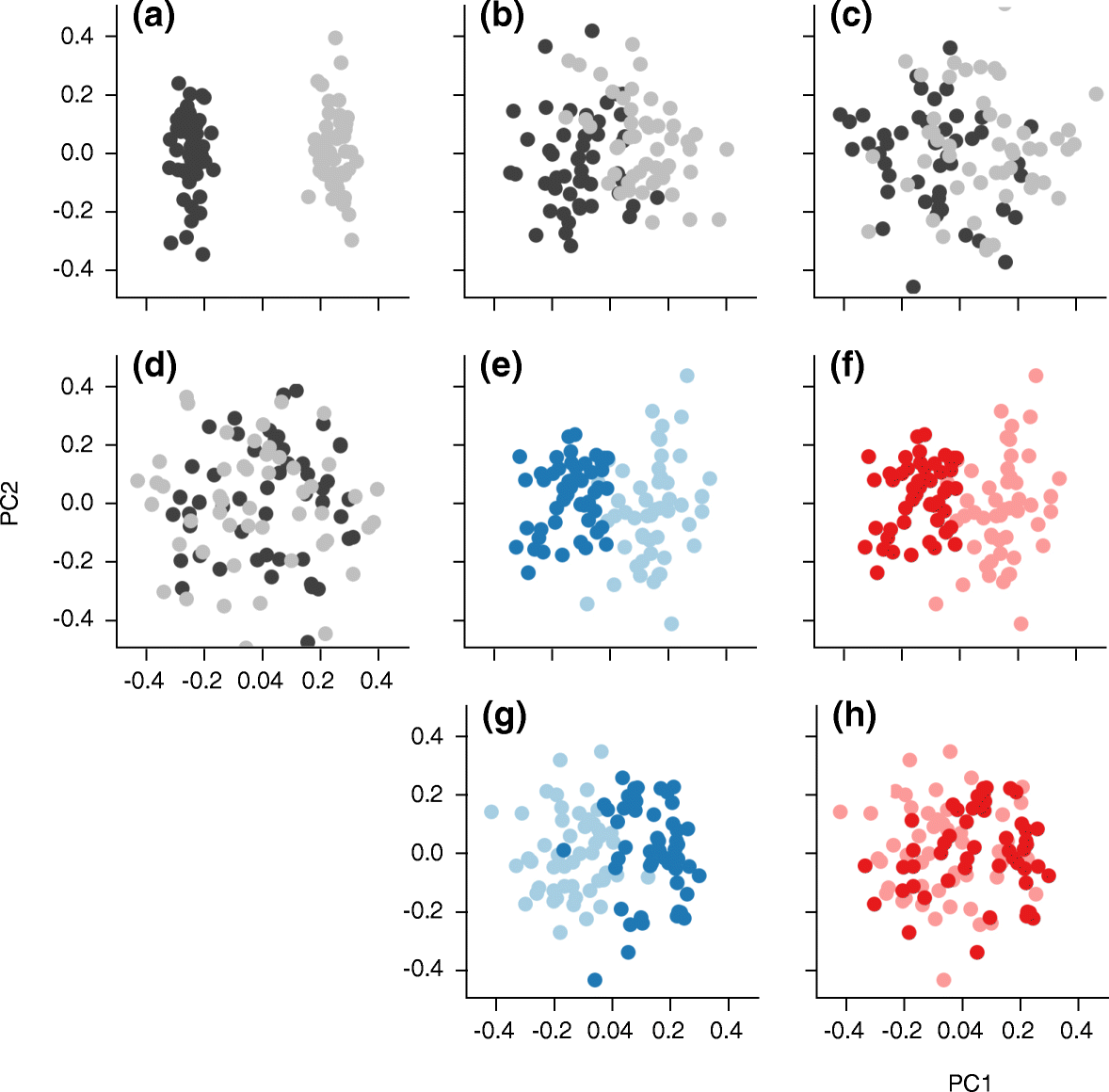
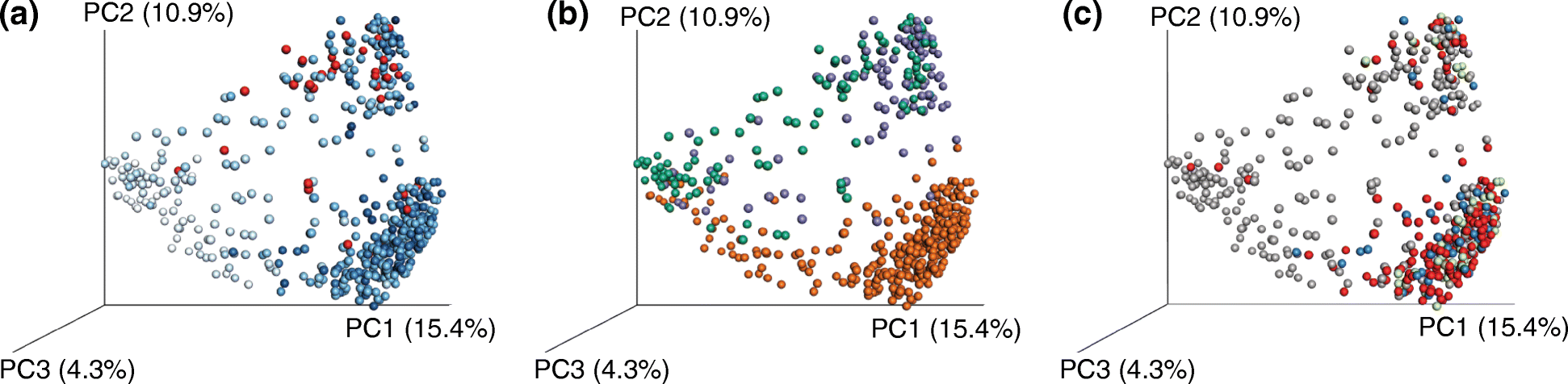
Similar articles
-
Adjusting for age improves identification of gut microbiome alterations in multiple diseases.Elife. 2020 Mar 11;9:e50240. doi: 10.7554/eLife.50240. Elife. 2020. PMID: 32159510 Free PMC article.
-
Subgroup analyses in randomised controlled trials: quantifying the risks of false-positives and false-negatives.Health Technol Assess. 2001;5(33):1-56. doi: 10.3310/hta5330. Health Technol Assess. 2001. PMID: 11701102 Review.
-
Ecophylogenetics Clarifies the Evolutionary Association between Mammals and Their Gut Microbiota.mBio. 2018 Sep 11;9(5):e01348-18. doi: 10.1128/mBio.01348-18. mBio. 2018. PMID: 30206171 Free PMC article.
-
Gut microbiome composition better reflects host phylogeny than diet diversity in breeding wood-warblers.Mol Ecol. 2023 Jan;32(2):518-536. doi: 10.1111/mec.16762. Epub 2022 Nov 20. Mol Ecol. 2023. PMID: 36325817
-
Characterization of the gut microbiome in epidemiologic studies: the multiethnic cohort experience.Ann Epidemiol. 2016 May;26(5):373-9. doi: 10.1016/j.annepidem.2016.02.009. Epub 2016 Mar 8. Ann Epidemiol. 2016. PMID: 27039047 Free PMC article. Review.
Cited by
-
A database of animal metagenomes.Sci Data. 2022 Jun 16;9(1):312. doi: 10.1038/s41597-022-01444-w. Sci Data. 2022. PMID: 35710683 Free PMC article.
-
Uniform Manifold Approximation and Projection (UMAP) Reveals Composite Patterns and Resolves Visualization Artifacts in Microbiome Data.mSystems. 2021 Oct 26;6(5):e0069121. doi: 10.1128/mSystems.00691-21. Epub 2021 Oct 5. mSystems. 2021. PMID: 34609167 Free PMC article.
-
Primer selection impacts the evaluation of microecological patterns in environmental microbiomes.Imeta. 2023 Sep 17;2(4):e135. doi: 10.1002/imt2.135. eCollection 2023 Nov. Imeta. 2023. PMID: 38868223 Free PMC article.
-
A comprehensive guide to assess gut mycobiome and its role in pathogenesis and treatment of inflammatory bowel disease.Indian J Gastroenterol. 2024 Feb;43(1):112-128. doi: 10.1007/s12664-023-01510-0. Epub 2024 Feb 27. Indian J Gastroenterol. 2024. PMID: 38409485 Review.
-
Oxidative stress in the oral cavity is driven by individual-specific bacterial communities.NPJ Biofilms Microbiomes. 2018 Nov 27;4:29. doi: 10.1038/s41522-018-0072-3. eCollection 2018. NPJ Biofilms Microbiomes. 2018. PMID: 30510769 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

