Alternative lengthening of human telomeres is a conservative DNA replication process with features of break-induced replication
- PMID: 27760777
- PMCID: PMC5167343
- DOI: 10.15252/embr.201643169
Alternative lengthening of human telomeres is a conservative DNA replication process with features of break-induced replication
Abstract
Human malignancies overcome replicative senescence either by activating the reverse-transcriptase telomerase or by utilizing a homologous recombination-based mechanism, referred to as alternative lengthening of telomeres (ALT). In budding yeast, ALT exhibits features of break-induced replication (BIR), a repair pathway for one-ended DNA double-strand breaks (DSBs) that requires the non-essential subunit Pol32 of DNA polymerase delta and leads to conservative DNA replication. Here, we examined whether ALT in human cancers also exhibits features of BIR A telomeric fluorescence in situ hybridization protocol involving three consecutive staining steps revealed the presence of conservatively replicated telomeric DNA in telomerase-negative cancer cells. Furthermore, depletion of PolD3 or PolD4, two subunits of human DNA polymerase delta that are essential for BIR, reduced the frequency of conservatively replicated telomeric DNA ends and led to shorter telomeres and chromosome end-to-end fusions. Taken together, these results suggest that BIR is associated with conservative DNA replication in human cells and mediates ALT in cancer.
Keywords: PolD3; PolD4; alternative lengthening of telomeres; break‐induced replication; telomere length regulation.
© 2016 The Authors. Published under the terms of the CC BY NC ND 4.0 license.
Figures

Semiconservative replication. Diagram of a chromosome with the telomeric C‐rich and G‐rich strands colored red and green, respectively. Newly synthesized strands are indicated by dotted lines. The three steps of the protocol were strand‐specific: dual color, denaturing FISH (1), non‐denaturing chromosome orientation (CO)‐FISH (2), and denaturing FISH (3). In the second step, the nascent strands have been digested. In all steps, two sets of PNA primers specific for the G‐strand and C‐strand, respectively, were used to monitor the presence of both strands. The arrows indicate the color of the emitted light and its intensity (idealized).
Conservative replication. Diagram showing that telomeric conservative replication (shown here to involve the entire length of the telomeres at the p arms) leads to distinct staining patterns, not observed with semiconservative replication. BIR, break‐induced replication.
Partially conservative and partially semiconservative telomeric replication. Diagram showing the staining patterns predicted for telomeres that are partially conservatively replicated (distal half) and partially semiconservatively replicated (proximal half), as might occur following fork collapse within a telomere (shown only for the telomeres at the p arms).
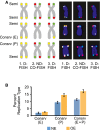
Examples of chromosome arms exhibiting features of telomeric semiconservative (Semi) replication, conservative (Consrv) replication of the entire telomere (E) and conservative replication of part of the telomere (P). Idealized chromosome diagrams (left) and actual microscopy images (right) are shown. D‐FISH, denaturing FISH; ND‐CO‐FISH, non‐denaturing CO‐FISH.
Percentages of chromosome arms exhibiting conservative (Consrv) replication of the entire telomere (E), conservative replication of part of the telomere (P), and conservative replication of either the entire telomere of part of it (E+P). NE, U2OS cells expressing normal levels of cyclin E; OE, U2OS cells overexpressing cyclin E for 4 days. Bars represent means and standard errors of the mean from three independent experiments. Cyclin E overexpression resulted in higher percentages of conservatively replicated telomeres: P < 0.025 for Consrv (P) and P < 0.05 for Consrv (E+P), as calculated using the paired t‐test.
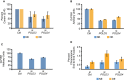
Decrease in the fraction of chromosome arms exhibiting conservative (Consrv) DNA replication of the entire telomere length or part of the telomere (E+P) in U2OS cells after depletion of POLD3 or POLD4 by siRNA. siCtrl, control siRNA; NE, cells expressing normal levels of cyclin E; OE, cells overexpressing cyclin E for 4 days. Bars represent means and standard errors of the mean from two independent experiments. For statistical comparisons, the effects of PolD3 and Pold4 depletion were examined after grouping together the data from the NE and OE cells. PolD3 depletion reduced the fraction of conservatively replicated telomeres at a level of P = 0.072, whereas for PolD4 depletion the decrease was significant at a level of P < 0.025, as calculated using the paired t‐test.
Decreased telomere length following PolD3 or PolD4 depletion in U2OS cells expressing normal levels of cyclin E (NE) or overexpressing cyclin E (OE). The cells were transfected with control siRNA (siCtrl) or siRNAs targeting POLD3 or POLD4 and, 3 days later, fixed and examined for telomere length by Q‐FISH. Bars indicate means and standard error of the mean from more than 2,600 telomeres examined per condition. The differences between control and PolD3‐ or PolD4‐depleted cells were significant (P < 10−6) for both NE and OE cells, as determined by unpaired t‐tests.
Decreased telomere length following PolD3 or PolD4 depletion in parental U2OS cells. The cells were transfected with control siRNA (siCtrl) or siRNAs targeting POLD3 or POLD4 and, 3 days later, fixed and examined for telomere length by Q‐FISH. Bars indicate means and standard error of the mean from more than 2,800 telomeres examined per condition. The differences between control and PolD3‐ or PolD4‐depleted cells were significant (P < 10−6), as determined by unpaired t‐tests.
Increased frequency of chromosome end‐to‐end fusions following POLD3 or POLD4 depletion. U2OS cells expressing normal levels of cyclin E (NE) or overexpressing cyclin E for 4 days (OE) were transfected with control siRNA (siCtrl) os siRNAs targeting POLD3 or POLD4. For each condition, 25 metaphases were examined (representing about 1,900 high‐quality chromosome arms per condition); the percentage of chromosome fusions was determined for each metaphase and used to calculate means and standard errors of the mean. For statistical analysis, the numbers of chromosome end‐to‐end fusions in the various conditions were compared by chi‐square tests. Cyclin E overexpression led to a higher number of fusions (P < 0.0005); PolD3 and PolD4 depletion also led to a higher number of fusions (P < 0.001 and P < 0.025, respectively).
References
-
- Harley CB, Futcher AB, Greider CW (1990) Telomeres shorten during ageing of human fibroblasts. Nature 345: 458–460 - PubMed
-
- Shay JW, Bacchetti S (1997) A survey of telomerase activity in human cancer. Eur J Cancer 33: 787–791 - PubMed
-
- Azzalin CM, Lingner J (2015) Telomere functions grounding on TERRA firma. Trends Cell Biol 25: 29–36 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

